Recently, a virus called SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) caused an epidemic with pneumonia as the main symptom. This SARS-CoV-2 is a coronavirus, so what exactly is a coronavirus?
Overview of Coronavirus
Coronavirus is a general term for a class of RNA viruses, belonging to the genus Nidovirales and the family Coronaviridae. The coronavirus has a characteristic crown-like appearance when observed under an electron microscope (Fig.1), hence the name.
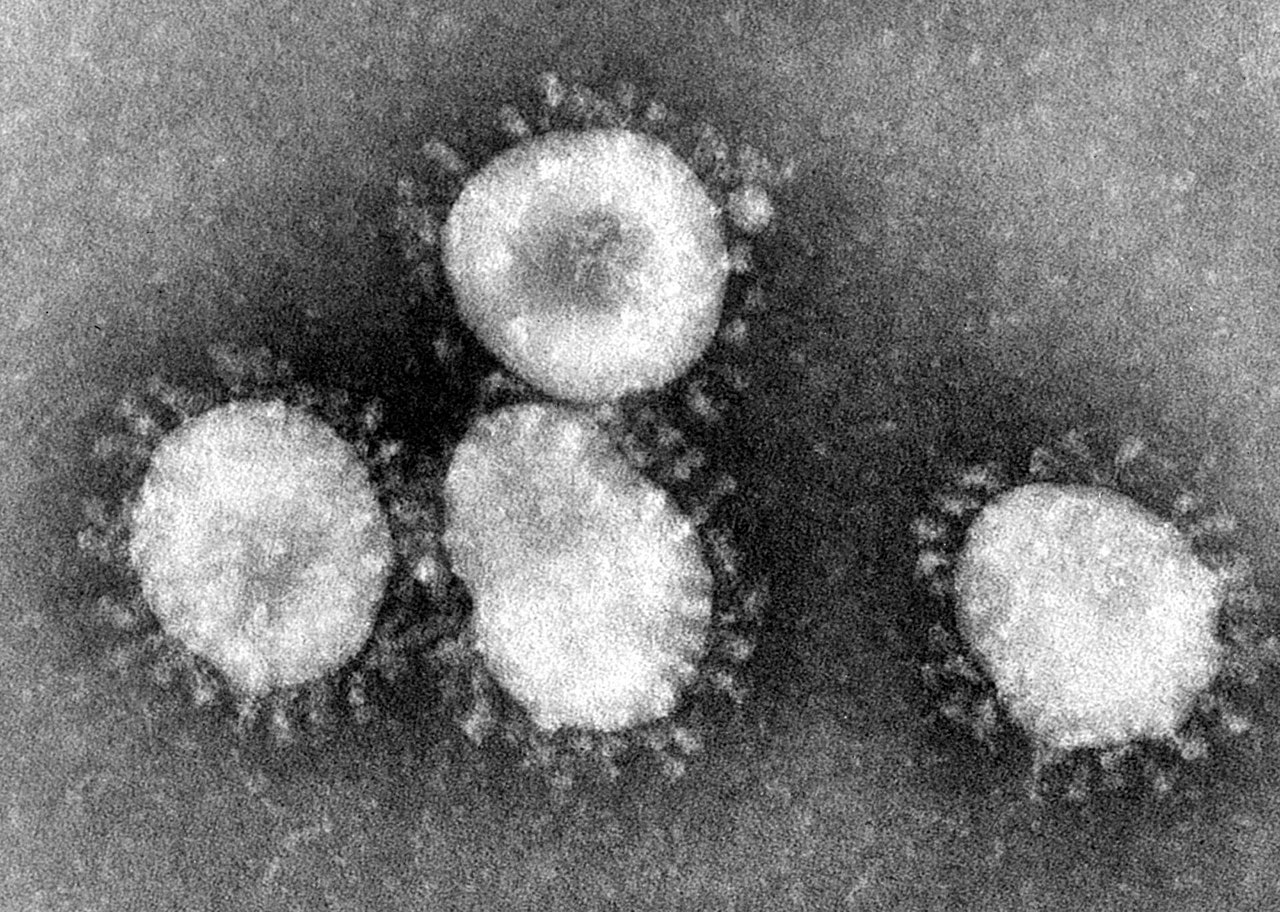
Fig.1 Morphology of Coronavirus particles as seen by electron microscopy
The coronavirus genome has a methylated cap structure at the 5’ end and a poly (A) tail at the 3’ end. The genome is a single-stranded positive-stranded RNA with a length of about 27-32kb, which is the largest genome of any RNA virus. Its virus particles are mostly spherical or oval and the virus has an envelope with spinous processes on it. Besides, the spinous processes of different coronaviruses are significantly different.
Coronavirus classification
Coronaviruses are classified according to the International Committee on Taxonomy of Viruses (ICTV). Coronaviruses can be divided into two subfamilies, Coronaviridae and Torovirinae. Coronavirus subfamilies are divided into α, β, γ, δ four genera. Among them, β-type coronavirus can be divided into four independent subgroups A, B, C and D (Fig. 2A).
There are currently six known coronaviruses that can infect humans in addition to 2019-nCoV. Among them, HCoV-229E and HCoV-NL63 belong to the alpha genus coronavirus, and HCoV-OC43, SARS-CoV, HCoV-HKU1 and MERS-CoV are all β genus coronaviruses. Among β-type coronaviruses, HCoV-OC43 and HCoV-HKU1 belong to subgroup A, SARS-CoV belongs to subgroup B, and MERS-CoV belongs to subgroup C (Fig. 2B). SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe infectious diseases in humans, while the other four viruses infect humans with milder symptoms.
Among other coronaviruses that infect animals, mammalian coronaviruses are mainly α and β coronaviruses, and avian coronaviruses are mainly derived from γ and δ coronaviruses (Fig. 2B).
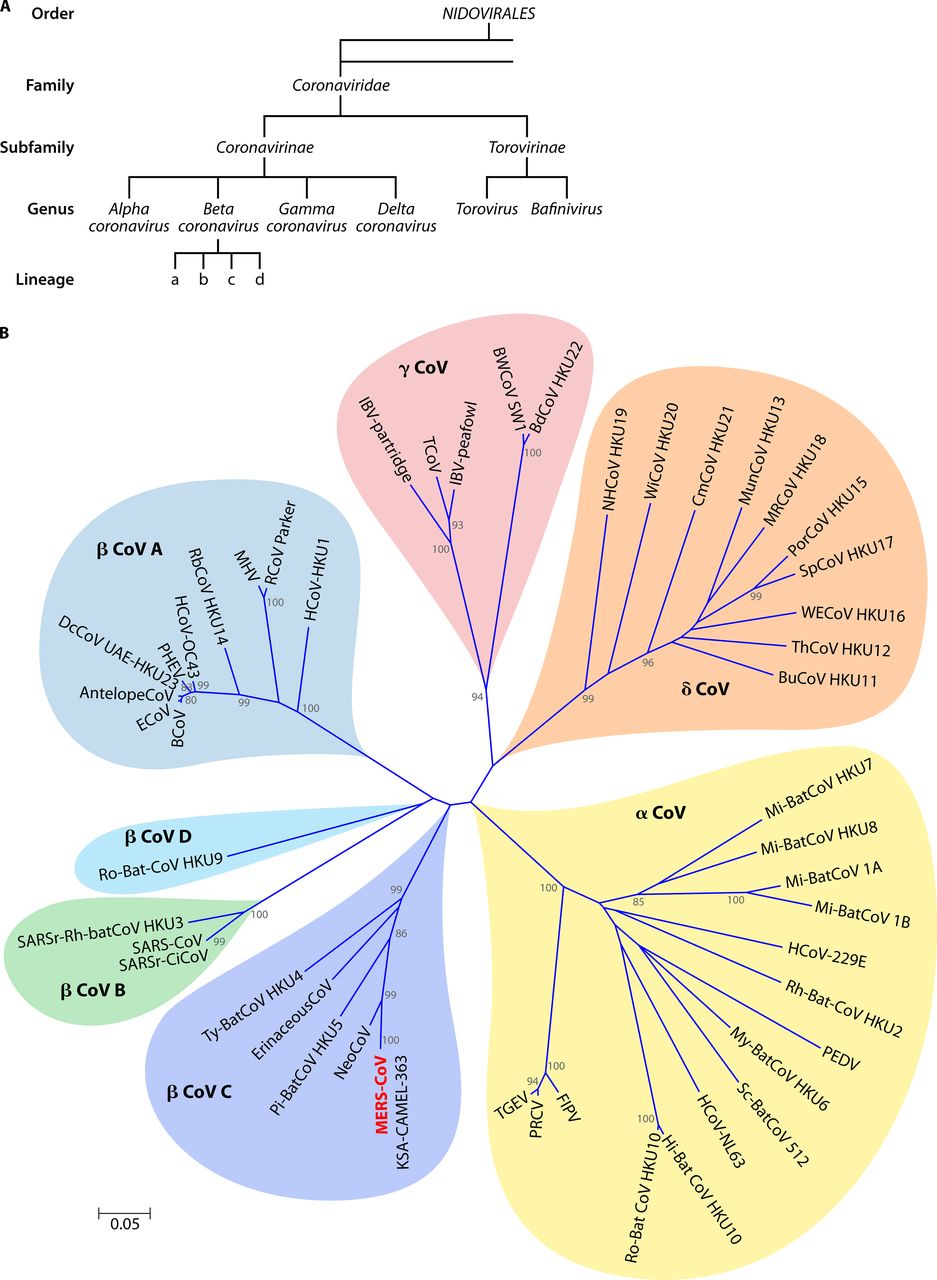
Fig.2 (A) The taxonomy of cornaviruses by ICTV. (B) Phylogenetic tree of 50 coronaviruses
Replication Cycle and Structure of Coronavirus
Coronaviruses attach to specific cellular receptors through the S protein, which triggers conformational changes in the S protein, and then mediates fusion between the virus membrane and the cell membrane, causing the nucleocapsid to be released into the cell. After entering the cell, the genomic RNA is translated and assembled through the host organelle, and mature virus particles are re-released out of the cell through the exocytosis of the host cell, and undergo a new round of replication cycle.
Coronavirus’ life cycle involves a variety of proteins, the most important of which are the following five structural proteins: spike (S) protein, membrane (M) protein, nucleocapsid (N) protein, hemagglutinin-esterase (HE) protein, and envelope (E) protein (Fig. 3).
- The S protein protrudes from the virus envelope and can form homotrimers, showing the coronal appearance of a coronavirus. It can mediate the binding and fusion of the virus with the host cell membrane through the receptor binding domain (RBD), so that the virus enters the cell.
- M protein is the most abundant virion membrane protein, which is responsible for the transmembrane transport of nutrients, the emergence of new virus buds, and the formation of virus outer membranes.
- N protein is related to the formation of nucleocapsid in the RNA genome and may be involved in regulating viral RNA synthesis and interact with M protein during virus budding.
- HE glycoproteins are only found in β coronavirus, HCoV-OC43 (Human Coronavirus OC43) and HKU1 (Human Coronavirus HKU1). The hemagglutinin moiety binds to neuraminic acid on the surface of the host cell, which may facilitate the virus’ initial adsorption of cell membranes.
- E protein can bind to the envelope, and its specific function is not clear. However, studies have confirmed that E protein, M and N proteins are necessary for SARS-CoV to correctly assemble and release the virus.
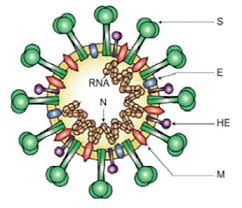
Fig.3 Coronavirus virion structure shown with structural proteins
Therapy of Coronavirus
These proteins play important roles in the various steps of the viral replication cycle, which can be effectively inhibited by antiviral drugs.
The development of existing anti-coronal virus drugs is mainly divided into non-specific anti-viral drugs and specific anti-viral drugs. Non-specific antiviral drugs include screening of existing antiviral drugs, molecules, and development of immunomodulators. The development of specific antiviral drugs has focused on host cell receptors or on coronavirus structural protein S, including vaccines, antisense nucleic acid drugs, and specific enzyme inhibitors.
Creative Biolabs is a leading service provider covering the entire antibody discovery and development process. We can provide you with coronavirus-related mouse/rabbit/human-derived recombinant antibodies, therapeutic antibodies and other related services to help your coronavirus drug development.
References:
[1] Weiss S R, Navasmartin S. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus[J]. Microbiology & Molecular Biology Reviews Mmbr, 2005, 69(4):635.
[2] Li F, Goff S P. Receptor Recognition Mechanisms of Coronaviruses: a Decade of Structural Studies[J]. Journal of Virology, 2015, 89(4):1954-1964.
[3] Tok T T, Tatar G. Structures and Functions of Coronavirus Proteins: Molecular Modeling of Viral Nucleoprotein.
