The paradigm of the adenoma-cancer sequence has typically focused on cancer cells in colorectal cancer (CRC) development that is already validated to entail heterologous interactions between altered cells and their surrounding microenvironment. To far, most research on advanced colorectal cancer has focused on bulk mapping to identify cancer driver genes and characterize biological pathways linked with cancer.
Researchers from Leiden University lately published an article in journal Gut entitled “Transcriptomic and immunophenotypic profiling reveals molecular and immunological hallmarks of colorectal cancer tumourigenesis” and identified biomarkers tightly associated with disease progression, as well as targeted immune processes that can be exploited in clinical settings.
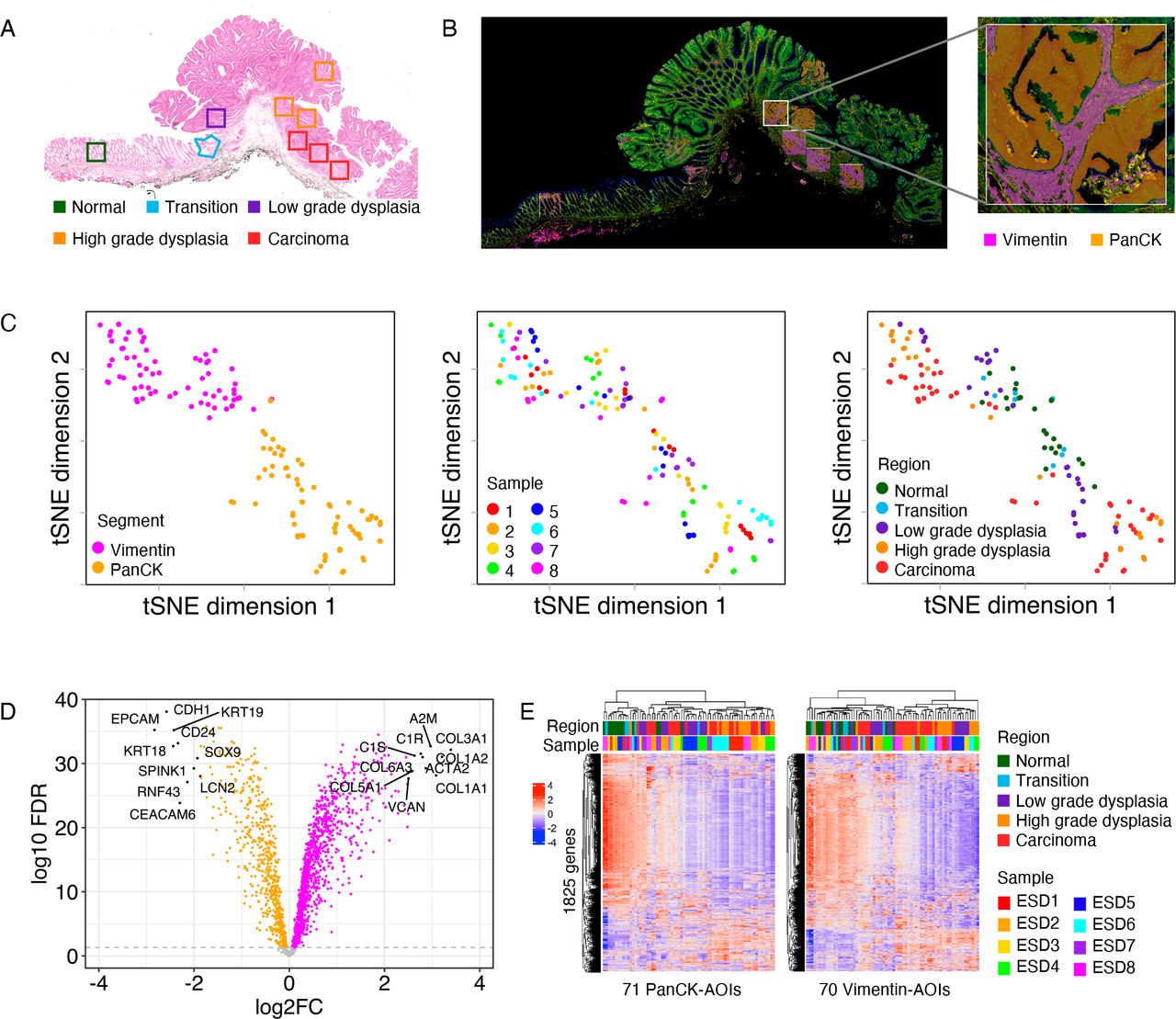
Biological insights into the progressive development and progression of colorectal cancer are essential to develop individualized early detection approaches and optimal clinical management measurements. In this study, the researchers’ goal was to dissect the transcriptional and immunological alterations that accompany colorectal cancer malignancy and to identify clinically relevant biomarkers through spatial mapping of PT1 colorectal cancer samples.
They used digital spatial mapping (GeoMx) on eight PT1 cancer tissues to study gene expression in epithelial and mesenchymal segments in different histological regions, including normal mucosa, low- and high-grade atypical hyperplasia, and carcinoma. Serial tissue sections were analyzed by imaging mass cytometry to depict the immune environment. Finally, publicly available single-cell RNA sequencing data were analyzed to determine the cellular origin of the transcripts of interest.
Comparison of gene expression in different regions of PT1 colorectal cancer samples revealed differentially expressed genes in epithelial (n=1394 genes) and mesenchymal segments (n=1145 genes) in different tissue structures. Pathway analysis revealed that inflammatory responses occurred earlier throughout malignant transformation, typically manifested by upregulation of gene signaling, such as innate immune sensing.
The researchers detected increased myeloid infiltration, macrophage transfer from normal tissue to cancer via dysplasia, and transfer from pro-inflammatory HLA-DR+CD204-macrophages to HLA-DR-CD204+ immunosuppressive subpopulations with upregulation of CD47/SIRPα “don’t-eat-me” signals.
In conclusion, this study demonstrates the use of digital spatial profiling in early colorectal cancer and provides insights into the biological mechanisms accompanying the colorectal cancer growth. The results suggest that innate immunity plays a critical role in colorectal carcinogenesis. In addition, the study’s methodology provides a framework for identifying biomarkers that vary throughout colorectal carcinogenesis. These insights pave the way for improved clinical care of patients with early colorectal cancer.
Reference
1. Roelands, Jessica, et al. “Transcriptomic and immunophenotypic profiling reveals molecular and immunological hallmarks of colorectal cancer tumourigenesis.” Gut 72.7 (2023): 1326-1339.