Cell Cycle
The cell cycle is an orchestrated sequence of events under very strict control, providing the fast to the slow development, growth, and maintenance of all living organisms. Basically, the cell cycle can be divided into interphase, which includes G1 (Gap 1), S (Synthesis), and G2 (Gap 2), and M phase (Mitosis), where the cell actually divides. Interphase: It forms a phase in the cell's life, and is majorly broken into the preparatory phase which entails cell growth in size, replication of DNA during the S phase, and preparation for mitosis in such a manner that all its components are rightly placed and duplicated in number. Being the climax of the cell cycle, mitosis is further broken down into prophase, metaphase, anaphase, and telophase until when the official cytoplasm division occurs and cells separate into daughter cells. The cell cycle is exceptionally complicated. It is regulated by a network of signaling pathways and checkpoints, which would obey just one thing: strict integrity of the genome and proper preparation of cells for the next phase. Errors in this regulation—the cell cycle—underlie the basis of cancer and the importance of the cell cycle in pathology.
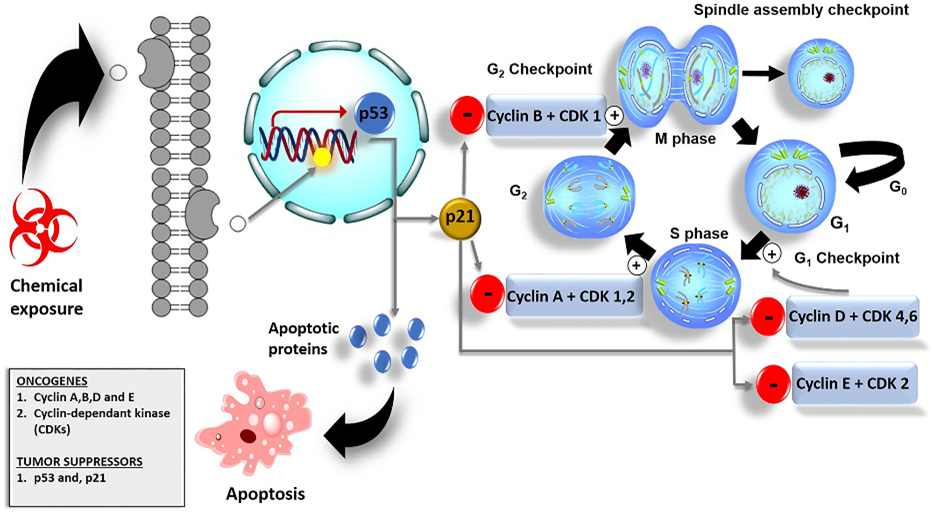 Figure 1 Cell cycle checkpoints and their role in cancer cell death. (Chota, 2021)
Figure 1 Cell cycle checkpoints and their role in cancer cell death. (Chota, 2021)
Cell Cycle Checkpoint
Cell cycle checkpoints are some of the surveillance mechanisms of the cell cycle deciding the progression after ensuring appropriate completion of respective the phase in order to take up the next. This sort of checkpoint is most preferably function to halt duplication with damaged DNA and maintain genomic stability without spreading errors to subsequent generations of cells. In principle, there are three kinds of checkpoints: the G1/S checkpoint, the G2/M checkpoint, and the spindle assembly checkpoint. The G1/S-checkpoint acts at the end of the G1 and at the beginning of the S-phase, the decision checkpoint of the cell that decides it to proceed for duplication of DNA or not. It through the checkpoints assesses the condition of the DNA integrity and availability of materials which the cell required synthesizing DNA. The G2/M-checkpoint ensures that all DNA is replicated, and if it is damaged in all, then the damaged part is repaired before it enters into division. Last in the line of cell cycle checkpoints is the spindle assembly checkpoint, where the cell is called upon to ensure that all the chromosomes are well attached to the spindle fibers before the division begins.
Cell Cycle Regulation
The progression in the cell cycle regulation is regulated with the help of different proteins which are involved as cyclins and cyclin-dependent kinases (CDKs). In the simplest form, cyclins are groupings of regulatory proteins that vary in amount at different times throughout the cycle of cells, and CDKs are enzymes that, after being activated by cyclins, phosphorylate proteins in the next phase of the cell cycle. The combination of specific cyclin-CDK complexes depends upon the time taken in the cell cycle and triggers further the events inside the cycle. Further tight control is subjected to such complexes by other processes that include synthesis and degradation of cyclin and regulation by phosphorylation and the action of CDK inhibitors. All processes leave the cell cycle in a position to dynamically respond to both internal and external signals, and thus they don't allow the signaling to move within a certain point.
DNA Repair
DNA repair mechanisms are required and important to correct mistakes occurring in DNA synthesis, to repair the damage induced either due to direct or indirect attack of environmental agents. In support of genetic information integrity nature has resulted in evolving many repair mechanisms which sulfide various types of DNA damage. End-independent productive They are base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), double-strand break (DSB) repair, homologous recombination (HR), and non-homologous end-joining (NHEJ). All these repair pathways include distinct subsets that will be recruited to repair specific defects—that is, specific types of damage. Similarly, BER will be responsible for fixing the small non-helix distorting base lesions. In the same breadth, while NER carries out the fixing of bulky helix-distorting lesions, MMR will fix replication errors such as base pairs, looped intermediaries created by insertion-deletions and other types of repair tolerable, include mechanisms for fixing DSBs. DSB mechanisms cover breaks in both strands of the DNA. DSB repair with HR is, for the most part, a function of the cell cycle S and G2 phases, since a sister chromatid, which is available for repair, and NHEJ, which does not require a template, is most active in G1.
p53 Pathway
The p53 pathway is one of the main key regulatory mechanisms of genomic stability in protection against cancer. For its potential to quench out cancerous cells, p53 is referred to as "guardian of the genome"; the protein is a very potent tumor suppressor protein as it involves intrinsic response towards damage at the level of DNA and towards stress signals in the cells. When operational, p53 is capable of mediating arrest of the cell cycle, permitting the repair of broken DNA, or eliciting apoptosis, programmed cell death when the damage is boundless and therefore in that instance it prevents the proliferation of broken cells. p53 acts by transcriptional control of a huge range of target genes involved in cell cycle arrest, repair of DNA, apoptosis and senescence. For example, p53 can activate the CDK inhibitor p21 for transcription activity that binds and inhibits cyclin-CDK complex and hence cause arrest. This then leaves time for the cell to repair its DNA before division. In case the damage is unrepairable, p53 can be used to initiate apoptosis to avoid the situation whereby damaged DNA gets to pass on to daughter cells.
- Chota, Alexander, Blassan P. George, and Heidi Abrahamse. "Interactions of multidomain pro-apoptotic and anti-apoptotic proteins in cancer cell death." Oncotarget 12.16 (2021): 1615. Distributed under Open Access license CC BY 4.0, without modification.
For research use only. Not intended for any clinical use.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.



