p53 Pathway
The p53 pathway is a cornerstone of cellular defense mechanisms against stress and damage, acting as a crucial regulator of cell fate in response to genomic instability, oncogene activation, and other stress signals. This tumor suppressor protein, often referred to as the "guardian of the genome," orchestrates a wide array of cellular responses, including cell cycle arrest, DNA repair, senescence, and apoptosis, thereby preventing the propagation of damaged DNA that could lead to tumorigenesis. The p53 protein functions primarily as a transcription factor, activating or repressing a multitude of target genes involved in various cellular processes critical for maintaining genomic integrity. Upon activation by cellular stress, p53 accumulates in the nucleus, where it can initiate a cell cycle pause, allowing for either repair of DNA damage or, if the damage is irreparable, the induction of programmed cell death. The pathway's importance is underscored by the fact that p53 is one of the most commonly mutated genes in human cancers, with mutations leading to loss of function and, consequently, uncontrolled cell growth and division. Moreover, the p53 pathway interacts with other signaling cascades, including those mediated by ATM and ATR kinases in response to DNA damage, highlighting its central role in the network of cellular stress responses. The pathway's ability to induce apoptosis in cells with excessive damage or oncogenic mutations serves as a critical barrier to cancer development, illustrating the indispensable role of p53 in tumor suppression and cellular homeostasis.
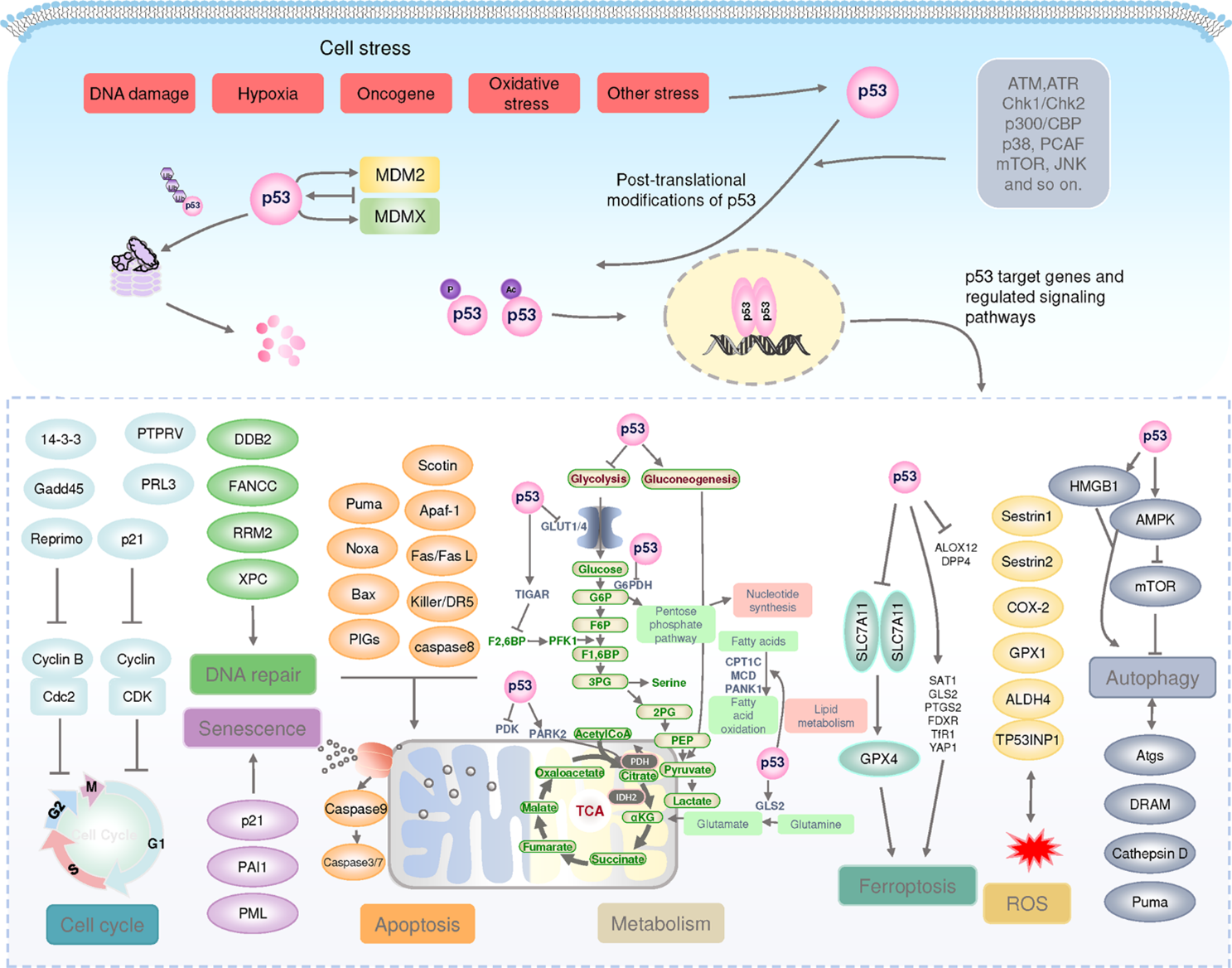 Figure 1 The p53 pathway. (Wang, 2023)
Figure 1 The p53 pathway. (Wang, 2023)
Representative Targets in p53 Pathway
p53
The p53 protein, encoded by the TP53 gene, stands as a cornerstone in the maintenance of genomic integrity and the suppression of tumorigenesis, meriting its designation as the "guardian of the genome." This multifaceted tumor suppressor plays a pivotal role in orchestrating a wide array of cellular responses to DNA damage, oxidative stress, and other oncogenic signals. By functioning as a transcription factor, p53 regulates the expression of a myriad of genes involved in DNA repair, cell cycle arrest, apoptosis, senescence, and metabolism, thus ensuring cellular adaptation or elimination in the face of genotoxic insults. The activation of p53 leads to cell cycle arrest in the G1 phase, allowing for DNA repair mechanisms to rectify damage or, in cases of irreparable damage, triggering apoptotic pathways to eliminate potentially malignant cells. Beyond these canonical roles, p53 also engages in modulating metabolic pathways and autophagy, highlighting its extensive influence on cellular homeostasis. Mutations in the TP53 gene, leading to the loss of normal p53 function, are among the most common genetic alterations observed in human cancers, underscoring the critical role of p53 in tumor prevention. The loss or inactivation of p53 not only contributes to the initiation and progression of cancer by permitting the survival and proliferation of cells with genomic aberrations but also influences the response of cancer cells to therapy.
NPM1
Nucleophosmin 1 (NPM1), also known as B23, numatrin, or NO38, is a multifunctional protein that plays critical roles in ribosome biogenesis, cell proliferation, and the stress response, positioning it as a pivotal player in maintaining cellular homeostasis and genomic integrity. Predominantly localized in the nucleolus, NPM1 is involved in several fundamental cellular processes, including the assembly and transport of ribosomes, the regulation of p53, and the response to DNA damage. It acts as a molecular chaperone, preventing protein aggregation and facilitating protein folding and transport within the nucleus. The ability of NPM1 to shuttle between the nucleus and cytoplasm further underscores its significance in various cellular functions, including the control of centrosome duplication and the regulation of ARF (alternate reading frame) tumor suppressor. Mutations and alterations in the expression of NPM1 have been implicated in the pathogenesis of several types of cancer, most notably acute myeloid leukemia (AML), where NPM1 mutations are among the most common genetic aberrations. These mutations typically lead to the aberrant cytoplasmic localization of NPM1, disrupting its normal functions and contributing to leukemogenesis through mechanisms that remain under investigation.



SIRT1
SIRT1, a member of the sirtuin family of NAD+-dependent deacetylases, is a pivotal enzyme implicated in the regulation of longevity, metabolism, and stress resistance. It functions by removing acetyl groups from various protein substrates, thereby modulating their activity, stability, and localization. This deacetylation process influences a wide array of cellular processes, including DNA repair, gene expression, mitochondrial biogenesis, and the cell cycle, positioning SIRT1 as a critical regulator of cellular homeostasis and survival. Through its interaction with key transcription factors and co-factors such as p53, FOXO proteins, and PGC-1α, SIRT1 integrates signals from the nutritional and external environment to adapt cellular physiology accordingly, promoting energy efficiency and stress resilience. Its role in deacetylating histones further contributes to the regulation of chromatin structure and the transcriptional silencing of genes, impacting aging and the development of age-related diseases. The upregulation of SIRT1 activity has been associated with increased lifespan in several model organisms, while its downregulation is linked to age-related pathologies, including cardiovascular diseases, neurodegenerative disorders, and various cancers. Consequently, SIRT1 has emerged as a promising target for therapeutic interventions aimed at ameliorating metabolic diseases, extending lifespan, and combating age-associated disorders.


Full List of Targets in p53 Pathway
| Biomarker | Alternative Names | Gene ID | UniProt ID | Roles |
| ATR | ATR Serine/Threonine Kinase; Ataxia Telangiectasia And Rad3-Related Protein; EC 2.7.11.1; FRP1; MEC1; MEC1, Mitosis Entry Checkpoint 1, Homolog; Ataxia Telangiectasia And Rad3 Related; Mitosis Entry Checkpoint 1; Homolog (S. Cerevisiae); FRAP-Related Protein-1; FRAP-Related Protein 1; FCTCS; SCKL1; SCKL | 545 | Q13535 | The protein encoded by this gene belongs the PI3/PI4-kinase family, and is most closely related to ATM, a protein kinase encoded by the gene mutated in ataxia telangiectasia. This protein and ATM share similarity with Schizosaccharomyces pombe rad3, a cell cycle checkpoint gene required for cell cycle arrest and DNA damage repair in response to DNA damage. This kinase has been shown to phosphorylate checkpoint kinase CHK1, checkpoint proteins RAD17, and RAD9, as well as tumor suppressor protein BRCA1. Mutations of this gene are associated with Seckel syndrome. An alternatively spliced transcript variant of this gene has been reported, however, its full length nature is not known. Transcript variants utilizing alternative polyA sites exist. |
| CLTC | Hc; CHC; CHC17; MRD56; CLH-17; CLTCL2 | 1213 | Q00610 | Clathrin is a major protein component of the cytoplasmic face of intracellular organelles, called coated vesicles and coated pits. These specialized organelles are involved in the intracellular trafficking of receptors and endocytosis of a variety of macromolecules. The basic subunit of the clathrin coat is composed of three heavy chains and three light chains. |
| CLTCL1 | Clathrin Heavy Chain Like 1; Clathrin Heavy Chain On Chromosome 22; Clathrin, Heavy Polypeptide-Like 1; CLH-22; CLTCL; CLH22; | 8218 | P53675 | This gene is a member of the clathrin heavy chain family and encodes a major protein of the polyhedral coat of coated pits and vesicles. Chromosomal aberrations involving this gene are associated with meningioma, DiGeorge syndrome, and velo-cardio-facial syndrome. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jun 2009] |
| CREBBP | CREBBP; CBP; CREB Binding Protein; KAT3A; RSTS; Rubinstein-Taybi Syndrome; CREB-Binding Protein | 1387 | Q92793 | This gene is ubiquitously expressed and is involved in the transcriptional coactivation of many different transcription factors. First isolated as a nuclear protein that binds to cAMP-response element binding protein (CREB), this gene is now known to play critical roles in embryonic development, growth control, and homeostasis by coupling chromatin remodeling to transcription factor recognition. The protein encoded by this gene has intrinsic histone acetyltransferase activity and also acts as a scaffold to stabilize additional protein interactions with the transcription complex. This protein acetylates both histone and non-histone proteins. This protein shares regions of very high sequence similarity with protein p300 in its bromodomain, cysteine-histidine-rich regions, and histone acetyltransferase domain. Mutations in this gene cause Rubinstein-Taybi syndrome (RTS). Chromosomal translocations involving this gene have been associated with acute myeloid leukemia. Alternative splicing results in multiple transcript variants encoding different isoforms. |
| EDA2R | EDA2R; ectodysplasin A2 receptor; XEDAR; EDAA2R; EDA-A2R; TNFRSF27 | 60401 | Q9HAV5 | The protein encoded by this gene is a type III transmembrane protein of the TNFR (tumor necrosis factor receptor) superfamily, and contains cysteine-rich repeats and a single transmembrane domain. This protein binds to the EDA-A2 isoform of ectodysplasin, which plays an important role in maintenance of hair and teeth. Alternatively spliced transcript variants encodes distinct protein isoforms. |
| EIF5A | Eukaryotic Translation Initiation Factor 5A; Rev-Binding Factor; EIF-5A-1; EIF-5A1; EIF-5A; EIF-4D | 1984 | P63241 | Enables U6 snRNA binding activity and protein N-terminus binding activity. Involved in several processes, including cellular response to virus; positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator; and tumor necrosis factor-mediated signaling pathway. Located in annulate lamellae; cytoplasm; and nucleus. Part of nuclear pore. |
| EP300 | E1A Binding Protein P300; EC 2.3.1.-; Protein Propionyltransferase P300; Histone Crotonyltransferase P300; Histone Butyryltransferase P300; Histone Acetyltransferase P300; E1A-Associated Protein P300 | 2033 | Q09472 | This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. It functions as histone acetyltransferase that regulates transcription via chromatin remodeling and is important in the processes of cell proliferation and differentiation. It mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. This gene has also been identified as a co-activator of HIF1A (hypoxia-inducible factor 1 alpha), and thus plays a role in the stimulation of hypoxia-induced genes such as VEGF. Defects in this gene are a cause of Rubinstein-Taybi syndrome and may also play a role in epithelial cancer. |
| IFITM2 | IFITM2; Interferon-Inducible Protein 1-8D; Dispanin Subfamily A Member 2c; DSPA2c; 1-8D; Interferon-Induced Transmembrane Protein 2; Interferon Induced Transmembrane Protein 2 (1-8D) | 10581 | Q01629 | IFN-induced antiviral protein which inhibits the entry of viruses to the host cell cytoplasm, permitting endocytosis, but preventing subsequent viral fusion and release of viral contents into the cytosol. Active against multiple viruses, including influenza A virus, SARS coronavirus (SARS-CoV), Marburg virus (MARV), Ebola virus (EBOV), Dengue virus (DNV), West Nile virus (WNV), human immunodeficiency virus type 1 (HIV-1) and vesicular stomatitis virus (VSV). Can inhibit: influenza virus hemagglutinin protein-mediated viral entry, MARV and EBOV GP1,2-mediated viral entry, SARS-CoV S protein-mediated viral entry and VSV G protein-mediated viral entry. Induces cell cycle arrest and mediates apoptosis by caspase activation and in p53-independent manner. |
| IFITM3 | Interferon Induced Transmembrane Protein 3 | 10410 | Q01628 | The protein encoded by this gene is an interferon-induced membrane protein that helps confer immunity to influenza A H1N1 virus, West Nile virus, and dengue virus. Two transcript variants, only one of them protein-coding, have been found for this gene. Another variant encoding an N-terminally truncated isoform has been reported, but the full-length nature of this variant has not been determined. [provided by RefSeq, May 2012] |
| ING1 | ING1; Inhibitor Of Growth 1; Growth Inhibitor ING1; P33; P47ING1a; Inhibitor Of Growth Family, Member 1; P33ING1b; Growth Inhibitory Protein ING1; Tumor Suppressor ING1; P24ING1c; P47 | 3621 | Q9UK53 | Cooperates with p53/TP53 in the negative regulatory pathway of cell growth by modulating p53-dependent transcriptional activation. Implicated as a tumor suppressor gene. |
| LRRC15 | LIB | 131578 | Q8TF66 | LRRC15 may play some role in innate immunity. LRRC15 is aberrantly expressed in cancer. It is highly expressed in CAFs within the stroma of numerous solid tumors and directly expressed in mesenchymal tumors such as glioblastoma, sarcomas, and melanoma. |
| MDM2 | MDM2 Proto-Oncogene; MDM2 Proto-Oncogene, E3 Ubiquitin Protein Ligase; Oncoprotein Mdm2; Hdm2; Mdm2, Transformed 3T3 Cell Double Minute 2, P53 Binding Protein (Mouse); Mdm2, Transformed 3T3 Cell Double Minute 2, P53 Binding Protein; Mouse Double Minute 2, Human Homolog Of; P53-Binding Protein; Double Minute 2, Human Homolog Of; P53-Binding Protein; Mdm2, P53 E3 Ubiquitin Protein Ligase Homolog; MDM2 Oncogene, E3 Ubiquitin Protein Ligase | 4193 | A7UKX8 | This gene encodes a nuclear-localized E3 ubiquitin ligase. The encoded protein can promote tumor formation by targeting tumor suppressor proteins, such as p53, for proteasomal degradation. This gene is itself transcriptionally-regulated by p53. Overexpression or amplification of this locus is detected in a variety of different cancers. There is a pseudogene for this gene on chromosome 2. Alternative splicing results in a multitude of transcript variants, many of which may be expressed only in tumor cells. [provided by RefSeq, Jun 2013] |
| NPM1 | Nucleophosmin 1; Nucleophosmin (Nucleolar Phosphoprotein B23, Numatrin); Nucleophosmin/Nucleoplasmin Family, Member 1; Nucleolar Phosphoprotein B23; Nucleolar Protein NO38; Nucleophosmin | 4869 | P06748 | The protein encoded by this gene is involved in several cellular processes, including centrosome duplication, protein chaperoning, and cell proliferation. The encoded phosphoprotein shuttles between the nucleolus, nucleus, and cytoplasm, chaperoning ribosomal proteins and core histones from the nucleus to the cytoplasm. This protein is also known to sequester the tumor suppressor ARF in the nucleolus, protecting it from degradation until it is needed. Mutations in this gene are associated with acute myeloid leukemia. Dozens of pseudogenes of this gene have been identified. [provided by RefSeq, Aug 2017] |
| NQO1 | NAD(P)H Quinone Dehydrogenase 1; Diaphorase (NADH/NADPH) (Cytochrome B-5 Reductase); NAD(P)H Dehydrogenase, Quinone 1; NAD(P)H:Quinone Oxidoreductase 1; Phylloquinone Reductase; Menadione Reductase; Quinone Reductase 1; DT-Diaphorase; Azoreductase; EC 1.6.5.2; NMOR1; DIA4 | 1728 | P15559 | This gene is a member of the NAD(P)H dehydrogenase (quinone) family and encodes a cytoplasmic 2-electron reductase. This FAD-binding protein forms homodimers and reduces quinones to hydroquinones. This protein's enzymatic activity prevents the one electron reduction of quinones that results in the production of radical species. Mutations in this gene have been associated with tardive dyskinesia (TD), an increased risk of hematotoxicity after exposure to benzene, and susceptibility to various forms of cancer. Altered expression of this protein has been seen in many tumors and is also associated with Alzheimer's disease (AD). Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008] |
| p53 | 7157 | K7PPA8 | ||
| SIRT1 | Sirtuin 1; Regulatory Protein SIR2 Homolog 1; SIR2-Like Protein 1; SIR2L1; Sirtuin (Silent Mating Type Information Regulation 2, S. Cerevisiae, Homolog) 1; Sirtuin (Silent Mating Type Information Regulation 2 Homolog) 1 (S. Cerevisiae); NAD-Dependent Protein Deacetylase Sirtuin-1 | 23411 | Q96EB6 | This gene encodes a member of the sirtuin family of proteins, homologs to the yeast Sir2 protein. Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes. The functions of human sirtuins have not yet been determined; however, yeast sirtuin proteins are known to regulate epigenetic gene silencing and suppress recombination of rDNA. Studies suggest that the human sirtuins may function as intracellular regulatory proteins with mono-ADP-ribosyltransferase activity. The protein encoded by this gene is included in class I of the sirtuin family. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2008] |
| STK11 | PJS; LKB1; hLKB1 | 6794 | Q15831 | The protein encoded by this gene is a serine/threonine kinase that regulates cell polarity and energy metabolism and functions as a tumor suppressor. Mutations in this gene have been associated with the autosomal dominant Peutz-Jeghers syndrome, as well as with skin, pancreatic, and testicular cancers. |
| TERF2 | Telomeric Repeat Binding Factor 2; TTAGGG Repeat-Binding Factor 2; Telomeric DNA-Binding Protein; TRBF2; TRF2; Telomeric Repeat Binding Protein 2; Telomeric Repeat-Binding Factor 2 | 7014 | Q15554 | This gene encodes a telomere specific protein, TERF2, which is a component of the telomere nucleoprotein complex. This protein is present at telomeres in metaphase of the cell cycle, is a second negative regulator of telomere length and plays a key role in the protective activity of telomeres. While having similar telomere binding activity and domain organization, TERF2 differs from TERF1 in that its N terminus is basic rather than acidic. |
| TFAP2C | ERF1; TFAP2G; hAP-2g; AP2-GAMMA | 7022 | Q92754 | The protein encoded by this gene is a sequence-specific DNA-binding transcription factor involved in the activation of several developmental genes. The encoded protein can act as either a homodimer or heterodimer with other family members and is induced during retinoic acid-mediated differentiation. |
| TIGAR | FR2BP; C12orf5 | 57103 | Q9NQ88 | This gene is regulated as part of the p53 tumor suppressor pathway and encodes a protein with sequence similarity to the bisphosphate domain of the glycolytic enzyme that degrades fructose-2,6-bisphosphate. The protein functions by blocking glycolysis and directing the pathway into the pentose phosphate shunt. Expression of this protein also protects cells from DNA damaging reactive oxygen species and provides some protection from DNA damage-induced apoptosis. The 12p13.32 region that includes this gene is paralogous to the 11q13.3 region. |
| TP53BP1 | Tumor Protein P53 Binding Protein 1; P53-Binding Protein 1; P53BP1; 53BP1; Tumor Suppressor P53-Binding Protein 1; Tumor Protein P53-Binding Protein, 1 | 7158 | Q12888 | This gene encodes a protein that functions in the DNA double-strand break repair pathway choice, promoting non-homologous end joining (NHEJ) pathways, and limiting homologous recombination. This protein plays multiple roles in the DNA damage response, including promoting checkpoint signaling following DNA damage, acting as a scaffold for recruitment of DNA damage response proteins to damaged chromatin, and promoting NHEJ pathways by limiting end resection following a double-strand break. These roles are also important during V(D)J recombination, class switch recombination and at unprotected telomeres. Alternative splicing results in multiple transcript variants encoding different isoforms. |
| UBE2A | UBC2; HHR6A; MRXSN; RAD6A; MRXS30 | 7319 | P49459 | The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, ubiquitin-conjugating enzymes, and ubiquitin-protein ligases. This gene encodes a member of the E2 ubiquitin-conjugating enzyme family. This enzyme is required for post-replicative DNA damage repair, and may play a role in transcriptional regulation. Mutations in this gene are associated with cognitive disability. Alternative splicing results in multiple transcript variants. |
| UBE2B | UBE2B; Ubiquitin-Conjugating Enzyme E2-17 KDa; RAD6B; E2-17kDa; Ubiquitin-Conjugating Enzyme E2B (RAD6 Homolog); RAD6 Homolog B; Ubiquitin Carrier Protein B; HR6B; EC 6.3.2.19; E2 Protein; HHR6B; Ubiquitin-Conjugating Enzyme E2 B; Ubiquitin-Conjugating En | 7320 | P63146 | The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, or E1s, ubiquitin-conjugating enzymes, or E2s, and ubiquitin-protein ligases, or E3s. This gene encodes a member of the E2 ubiquitin-conjugating enzyme family. This enzyme is required for post-replicative DNA damage repair. Its protein sequence is 100% identical to the mouse, rat, and rabbit homologs, which indicates that this enzyme is highly conserved in eukaryotic evolution. |
| USP4 | USP4; Ubiquitin Thiolesterase 4; Ubiquitin Carboxyl-Terminal Esterase 4; Ubiquitin Carboxyl-Terminal Hydrolase 4; EC 3.4.19.12; Ubiquitin Thioesterase 4; Deubiquitinating Enzyme 4; Ubiquitin Specific Peptidase 4 (Proto-Oncogene); Ubiquitin Specific Protea | 7375 | Q13107 | Hydrolase that deubiquitinates target proteins such as the receptor ADORA2A, PDPK1 and TRIM21. Deubiquitination of ADORA2A increases the amount of functional receptor at the cell surface. May regulate mRNA splicing through deubiquitination of the U4 spliceosomal protein PRPF3. This may prevent its recognition by the U5 component PRPF8 thereby destabilizing interactions within the U4/U6.U5 snRNP. May also play a role in the regulation of quality control in the ER. |
| USP7 | Ubiquitin Specific Peptidase 7; Ubiquitin Specific Peptidase 7 (Herpes Virus-Associated); Ubiquitin-Specific-Processing Protease 7; Deubiquitinating Enzyme 7; Ubiquitin Thioesterase 7; HAUSP; Ubiquitin Specific Protease 7 (Herpes Virus-Associated) | 7874 | B7ZAX6 | The protein encoded by this gene belongs to the peptidase C19 family, which includes ubiquitinyl hydrolases. This protein deubiquitinates target proteins such as p53 (a tumor suppressor protein) and WASH (essential for endosomal protein recycling), and regulates their activities by counteracting the opposing ubiquitin ligase activity of proteins such as HDM2 and TRIM27, involved in the respective process. Mutations in this gene have been implicated in a neurodevelopmental disorder. [provided by RefSeq, Mar 2016] |
| VRK1 | PCH1; PCH1A | 7443 | Q99986 | This gene encodes a member of the vaccinia-related kinase (VRK) family of serine/threonine protein kinases. This gene is widely expressed in human tissues and has increased expression in actively dividing cells, such as those in testis, thymus, fetal liver, and carcinomas. Its protein localizes to the nucleus and has been shown to promote the stability and nuclear accumulation of a transcriptionally active p53 molecule and, in vitro, to phosphorylate Thr18 of p53 and reduce p53 ubiquitination. This gene, therefore, may regulate cell proliferation. This protein also phosphorylates histone, casein, and the transcription factors ATF2 (activating transcription factor 2) and c-JUN. [provided by RefSeq, Jul 2008] |
| WISP1 | WISP1; WNT1 inducible signaling pathway protein 1; CCN4; WISP1c; WISP1i; WISP1tc | 8840 | O95388 | This gene encodes a member+AK267:AL268 of the WNT1 inducible signaling pathway (WISP) protein subfamily, which belongs to the connective tissue growth factor (CTGF) family. WNT1 is a member of a family of cysteine-rich, glycosylated signaling proteins that mediate diverse developmental processes. The CTGF family members are characterized by four conserved cysteine-rich domains: insulin-like growth factor-binding domain, von Willebrand factor type C module, thrombospondin domain and C-terminal cystine knot-like domain. This gene may be downstream in the WNT1 signaling pathway that is relevant to malignant transformation. It is expressed at a high level in fibroblast cells, and overexpressed in colon tumors. The encoded protein binds to decorin and biglycan, two members of a family of small leucine-rich proteoglycans present in the extracellular matrix of connective tissue, and possibly prevents the inhibitory activity of decorin and biglycan in tumor cell proliferation. It also attenuates p53-mediated apoptosis in response to DNA damage through activation of the Akt kinase. It is 83% identical to the mouse protein at the amino acid level. Multiple alternatively spliced transcript variants have been identified. |
Tested Data-Supported Products for Targeting p53 Pathway
- Wang, Haolan, et al. "Targeting p53 pathways: mechanisms, structures, and advances in therapy." Signal transduction and targeted therapy 8.1 (2023): 92. Distributed under Open Access license CC BY 4.0, without modification.
For research use only. Not intended for any clinical use.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.



























