Apoptosis Transcription Factors and Regulators
Apoptosis transcription factors and regulators are essential in modulating the gene expression profiles that drive programmed cell death. These transcriptional regulators ensure that apoptosis is executed in a controlled and precise manner, responding appropriately to cellular stress and damage signals.
One of the central transcription factors in the regulation of apoptosis is p53, often referred to as the "guardian of the genome." This protein is pivotal in responding to a variety of stress signals, including DNA damage, oxidative stress, and oncogene activation. Upon activation, p53 can trigger the transcription of numerous pro-apoptotic genes, such as Bax, Noxa, and PUMA, which promote mitochondrial outer membrane permeabilization and the release of apoptotic factors. p53 also influences apoptosis indirectly by repressing the transcription of anti-apoptotic proteins like Bcl-2. The regulatory pathways involving p53 are critical in cancer biology, as mutations in the TP53 gene, which encodes p53, are highly prevalent in human cancers, often leading to the evasion of apoptosis and unchecked cellular proliferation.
Another important group of transcription factors involved in apoptosis are those belonging to the NF-κB family. Typically associated with promoting survival and inflammation, NF-κB can also have dual roles in apoptosis regulation, depending on the cellular context and the nature of the apoptotic signals. Under certain conditions, NF-κB activation can lead to the induction of anti-apoptotic genes, providing a survival advantage. However, in other contexts, particularly in response to specific cytokines or immune responses, NF-κB can promote apoptotic pathways.
In addition, the FOXO family of transcription factors, particularly FOXO3, plays a significant role in apoptosis. Under stress conditions, such as oxidative stress or nutrient deprivation, FOXO3 is activated and can move into the nucleus, where it upregulates the expression of genes involved in cell cycle arrest and apoptosis, including the Fas ligand – a critical component of the extrinsic apoptotic pathway.
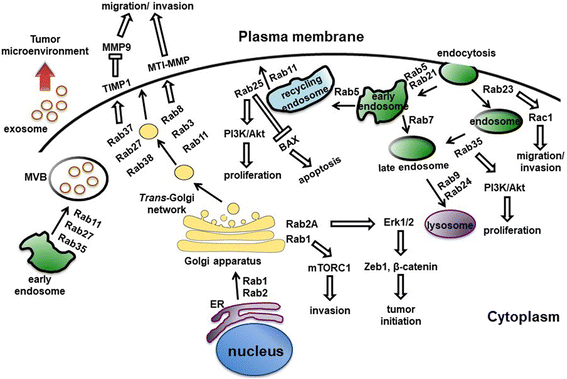 Figure 1 Rab proteins-mediated vesicular transport and signaling pathways. (Tzeng, 2016)
Figure 1 Rab proteins-mediated vesicular transport and signaling pathways. (Tzeng, 2016)
Representative Apoptosis Transcription Factors and Regulators
STAT1
STAT1, or Signal Transducer and Activator of Transcription 1, is a transcription factor that plays a central role in mediating cellular responses to interferons (IFNs), a group of cytokines involved in the immune response to viral and bacterial infections, as well as other immune and inflammatory stimuli. Upon activation by phosphorylation, typically by JAK kinases in response to IFN signaling, STAT1 forms homodimers or heterodimers with other STAT proteins and translocates to the nucleus, where it regulates the expression of target genes by binding to specific DNA sequences known as gamma-activated sequences (GAS). STAT1 is a key regulator of the innate and adaptive immune responses, influencing the expression of genes involved in anti-viral defense, inflammation, and immune modulation. It plays a critical role in activating immune cells such as macrophages, dendritic cells, and T cells, enabling them to mount an effective response against pathogens. Additionally, STAT1 is involved in the regulation of cell growth, apoptosis, and tumor surveillance, contributing to its function as a tumor suppressor. Dysregulation of STAT1 signaling has been implicated in various diseases, including infectious diseases, autoimmune disorders, and cancer. In some cases, mutations or deficiencies in STAT1 can lead to immunodeficiency, making individuals more susceptible to infections. Conversely, hyperactivation of STAT1 is associated with chronic inflammation and autoimmune conditions, such as lupus and multiple sclerosis. Moreover, alterations in STAT1 expression or activity are observed in many types of cancer, where it can either promote or suppress tumor growth depending on the cellular context.



STAT3
STAT3, or Signal Transducer and Activator of Transcription 3, is another member of the STAT protein family that plays a critical role in cellular signaling and gene regulation. Activated in response to cytokines and growth factors, STAT3 is involved in various cellular processes including cell growth, survival, proliferation, and differentiation. This transcription factor becomes activated through phosphorylation by receptor-associated kinases like JAK (Janus kinase), after which it dimerizes and translocates to the nucleus to induce the transcription of target genes. STAT3 is particularly noteworthy for its dual roles in both the immune system and cancer. In the immune system, STAT3 mediates the effects of interleukins, such as IL-6 and IL-10, which are crucial for the inflammatory response and immune regulation. In cancer, STAT3 is frequently hyperactivated, and its persistent activation is associated with oncogenesis and cancer progression. It promotes oncogenic processes by upregulating genes involved in cell cycle progression and survival, angiogenesis, and metastasis, and by suppressing anti-tumor immune responses. The involvement of STAT3 in so many critical pathways makes it a significant target for therapeutic intervention. In cancer, for instance, inhibitors of STAT3 are considered potential treatments because they can block the tumor-promoting actions of STAT3. Likewise, due to its role in inflammation and autoimmunity, targeting STAT3 could help manage diseases characterized by overactive immune responses, such as rheumatoid arthritis and other inflammatory conditions.



RELA
RELA, also known as p65, is a crucial component of the NF-kappaB (nuclear factor kappa-light-chain-enhancer of activated B cells) transcription factor complex, which plays a pivotal role in regulating immune responses, inflammation, and cellular survival. RELA specifically refers to one of the most studied subunits of the NF-kappaB complex, which is essential for NF-kappaB's transcriptional activity and its ability to induce gene expression in response to various stimuli, including cytokines, free radicals, ultraviolet irradiation, and bacterial or viral antigens. Upon activation, RELA translocates to the nucleus as part of the NF-kappaB heterodimer, typically pairing with another protein such as p50. In the nucleus, RELA binds to specific DNA sequences to regulate the transcription of genes involved in immune and inflammatory responses, such as cytokines, chemokines, and adhesion molecules. Additionally, RELA is involved in the control of genes that regulate cell proliferation and apoptosis, making it integral not only to immune function and inflammation but also to cell growth and survival.
The dysregulation of RELA and the NF-kappaB pathway is linked to a multitude of diseases, including inflammatory disorders, autoimmune diseases, and various types of cancer. In cancer, for example, persistent activation of RELA can lead to increased cell proliferation, survival, and angiogenesis, all of which support tumor growth and resistance to chemotherapy. In inflammatory diseases, such as rheumatoid arthritis and inflammatory bowel disease, aberrant RELA activity contributes to chronic inflammation and tissue damage.



Full List of Apoptosis Transcription Factors and Regulators
| Biomarker | Alternative Names | Gene ID | UniProt ID | Roles |
| C1D | C1D Nuclear Receptor Corepressor; Small Unique Nuclear Receptor Co-Repressor; C1D Nuclear Receptor Co-Repressor; HC1D; Small Unique Nuclear Receptor Corepressor; Nuclear DNA-Binding Protein; C1D DNA-Binding Protein; SUN-CoR; SUNCOR; Rrp47; LRP1 | 10438 | Q13901 | The protein encoded by this gene is a DNA binding and apoptosis-inducing protein and is localized in the nucleus. It is also a Rac3-interacting protein which acts as a corepressor for the thyroid hormone receptor. This protein is thought to regulate TRAX/Translin complex formation. Alternate splicing results in multiple transcript variants that encode the same protein. Multiple pseudogenes of this gene are found on chromosome 10. |
| CITED2 | ASD8; MRG1; VSD2; MRG-1; P35SRJ | 10370 | Q99967 | The protein encoded by this gene inhibits transactivation of HIF1A-induced genes by competing with binding of hypoxia-inducible factor 1-alpha to p300-CH1. Mutations in this gene are a cause of cardiac septal defects. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene. |
| c-Myc | MYC Proto-Oncogene, BHLH Transcription Factor; V-Myc Avian Myelocytomatosis Viral Oncogene Homolog; Class E Basic Helix-Loop-Helix Protein 39; Transcription Factor P64; Proto-Oncogene C-Myc; BHLHe39; Myc-Related Translation/Localization Regulatory Factor | 4609 | P01106 | This gene is a proto-oncogene and encodes a nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation. The encoded protein forms a heterodimer with the related transcription factor MAX. This complex binds to the E box DNA consensus sequence and regulates the transcription of specific target genes. Amplification of this gene is frequently observed in numerous human cancers. Translocations involving this gene are associated with Burkitt lymphoma and multiple myeloma in human patients. There is evidence to show that translation initiates both from an upstream, in-frame non-AUG (CUG) and a downstream AUG start site, resulting in the production of two isoforms with distinct N-termini. [provided by RefSeq, Aug 2017] |
| E2F1 | E2F1; RBBP3; Transcription factor E2F1; E2F-1; PBR3; Retinoblastoma-associated protein 1; RBAP-1; Retinoblastoma-binding protein 3; RBBP-3; pRB-binding protein E2F-1 | 1869 | Q01094 | The protein encoded by this gene is a member of the E2F family of transcription factors. The E2F family plays a crucial role in the control of cell cycle and action of tumor suppressor proteins and is also a target of the transforming proteins of small DNA tumor viruses. |
| E2F2 | E2F-2 | 1870 | Q14209 | The protein encoded by this gene is a member of the E2F family of transcription factors. The E2F family plays a crucial role in the control of cell cycle and action of tumor suppressor proteins and is also a target of the transforming proteins of small DNA tumor viruses. The E2F proteins contain several evolutionally conserved domains found in most members of the family. These domains include a DNA binding domain, a dimerization domain which determines interaction with the differentiation regulated transcription factor proteins (DP), a transactivation domain enriched in acidic amino acids, and a tumor suppressor protein association domain which is embedded within the transactivation domain. This protein and another 2 members, E2F1 and E2F3, have an additional cyclin binding domain. This protein binds specifically to retinoblastoma protein pRB in a cell-cycle dependent manner, and it exhibits overall 46% amino acid identity to E2F1. |
| E2F4 | E2F4 | 1874 | Q16254 | Transcription factor E2F4 is a protein that in humans is encoded by the E2F4 gene. |
| ETS1 | ETS Proto-Oncogene 1, Transcription Factor; Avian Erythroblastosis Virus E26 (V-Ets) Oncogene Homolog-1; V-Ets Avian Erythroblastosis Virus E26 Oncogene Homolog 1; EWSR2; P54; V-Ets Avian Erythroblastosis Virus E2 Oncogene Homolog 1 | 2113 | P14921 | This gene encodes a member of the ETS family of transcription factors, which are defined by the presence of a conserved ETS DNA-binding domain that recognizes the core consensus DNA sequence GGAA/T in target genes. These proteins function either as transcriptional activators or repressors of numerous genes, and are involved in stem cell development, cell senescence and death, and tumorigenesis. Alternatively spliced transcript variants encoding different isoforms have been described for this gene.[provided by RefSeq, Jul 2011] |
| FOS | c-fos | 2353 | P01100 | Enables DNA-binding transcription factor activity; double-stranded DNA binding activity; and sequence-specific DNA binding activity. Involved in several processes, including conditioned taste aversion; positive regulation of pri-miRNA transcription by RNA polymerase II; and response to steroid hormone. Located in membrane; neuron projection; and nucleus. Biomarker of congestive heart failure; glomerulonephritis; and hypertension. Orthologous to human FOS (Fos proto-oncogene, AP-1 transcription factor subunit). |
| FOSB | AP-1; G0S3; GOS3; GOSB | 2354 | P53539 | The Fos gene family consists of 4 members: FOS, FOSB, FOSL1, and FOSL2. These genes encode leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. As such, the FOS proteins have been implicated as regulators of cell proliferation, differentiation, and transformation. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. |
| FOXO3 | FOXO2; AF6q21; FKHRL1; FOXO3A; FKHRL1P2 | 2309 | O43524 | This gene belongs to the forkhead family of transcription factors which are characterized by a distinct forkhead domain. This gene likely functions as a trigger for apoptosis through expression of genes necessary for cell death. Translocation of this gene with the MLL gene is associated with secondary acute leukemia. Alternatively spliced transcript variants encoding the same protein have been observed. |
| IRF6 | Interferon Regulatory Factor 6 | 3664 | O14896 | This gene encodes a member of the interferon regulatory transcription factor (IRF) family. Family members share a highly-conserved N-terminal helix-turn-helix DNA-binding domain and a less conserved C-terminal protein-binding domain. The encoded protein may be a transcriptional activator. Mutations in this gene can cause van der Woude syndrome and popliteal pterygium syndrome. Mutations in this gene are also associated with non-syndromic orofacial cleft type 6. Alternate splicing results in multiple transcript variants. |
| IRF8 | Interferon Regulatory Factor 8 | 3394 | Q02556 | Interferon consensus sequence-binding protein (ICSBP) is a transcription factor of the interferon (IFN) regulatory factor (IRF) family. Proteins of this family are composed of a conserved DNA-binding domain in the N-terminal region and a divergent C-terminal region that serves as the regulatory domain. The IRF family proteins bind to the IFN-stimulated response element (ISRE) and regulate expression of genes stimulated by type I IFNs, namely IFN-alpha and IFN-beta. IRF family proteins also control expression of IFN-alpha and IFN-beta-regulated genes that are induced by viral infection. |
| JUND | JunD Proto-Oncogene, AP-1 Transcription Factor Subunit; Transcription Factor Jun-D; Activator Protein 1; JunD-FL Isoform; Jun D Proto-Oncogene; AP-1 | 3727 | P17535 | The protein encoded by this intronless gene is a member of the JUN family, and a functional component of the AP1 transcription factor complex. This protein has been proposed to protect cells from p53-dependent senescence and apoptosis. Alternative translation initiation site usage results in the production of different isoforms (PMID:12105216). [provided by RefSeq, Nov 2013]JUND (JunD Proto-Oncogene, AP-1 Transcription Factor Subunit) is a Protein Coding gene. Diseases associated with JUND include T-Cell Leukemia and Adult T-Cell Leukemia. Among its related pathways are ERK Signaling and EGF/EGFR Signaling Pathway. Gene Ontology (GO) annotations related to this gene include DNA binding transcription factor activity and enzyme binding. An important paralog of this gene is JUN. |
| MAD1L1 | MAD1; PIG9; TP53I9; TXBP181; mitotic spindle assembly checkpoint protein MAD1; MAD1 mitotic arrest deficient like 1; MAD1-like protein 1; mitotic arrest deficient 1-like protein 1; mitotic checkpoint MAD1 protein homolog; mitotic-arrest deficient 1, yeast, homolog-like 1; tax-binding protein 181; tumor protein p53 inducible protein 9 | 8379 | Q9Y6D9 | MAD1L1 is a component of the mitotic spindle-assembly checkpoint that prevents the onset of anaphase until all chromosome are properly aligned at the metaphase plate. MAD1L1 functions as a homodimer and interacts with MAD2L1. MAD1L1 may play a role in cell cycle control and tumor suppression. |
| MAF | CCA4; AYGRP; c-MAF; CTRCT21 | 4094 | O75444 | The protein encoded by this gene is a DNA-binding, leucine zipper-containing transcription factor that acts as a homodimer or as a heterodimer. Depending on the binding site and binding partner, the encoded protein can be a transcriptional activator or repressor. This protein plays a role in the regulation of several cellular processes, including embryonic lens fiber cell development, increased T-cell susceptibility to apoptosis, and chondrocyte terminal differentiation. Defects in this gene are a cause of juvenile-onset pulverulent cataract as well as congenital cerulean cataract 4 (CCA4). Two transcript variants encoding different isoforms have been found for this gene. |
| MAX | bHLHd4 | 4149 | P61244 | MYC associated factor X contains 1 basic helix-loop-helix (bHLH) domain and belongs to MAX family. It is highly expressed in the brain, heart and lung while lower levels are seen in the liver, kidney and skeletal muscle. MYC associated factor X can form homodimers and heterodimers with other family members, which include Mad, Mxi1 and Myc. Myc is an oncoprotein implicated in cell proliferation, differentiation and apoptosis. The homodimers and heterodimers compete for a common DNA target site (the E box) and rearrangement among these dimer forms provides a complex system of transcriptional regulation. MYC associated factor X may also repress transcription via the recruitment of a chromatin remodeling complex containing H3 Lys-9 histone methyltransferase activity. Multiple alternatively spliced transcript variants have been described for MYC associated factor X gene but the full-length nature for some of them is unknown. |
| MSX1 | Msh Homeobox 1; Msh Homeobox 1-Like Protein; Homeobox Protein Hox-7; HOX7; Msh (Drosophila) Homeo Box Homolog 1 (Formerly Homeo Box 7); Msh Homeobox Homolog 1 (Drosophila); Homeobox Protein MSX-1 | 17701 | P13297 | This gene encodes a member of the muscle segment homeobox gene family. The encoded protein functions as a transcriptional repressor during embryogenesis through interactions with components of the core transcription complex and other homeoproteins. It may also have roles in limb-pattern formation, craniofacial development, particularly odontogenesis, and tumor growth inhibition. Mutations in this gene, which was once known as homeobox 7, have been associated with nonsyndromic cleft lip with or without cleft palate 5, Witkop syndrome, Wolf-Hirschom syndrome, and autosomoal dominant hypodontia. |
| MYC | MRTL; MYCC; c-Myc; bHLHe39 | 4609 | P01106 | This gene is a proto-oncogene and encodes a nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis and cellular transformation. The encoded protein forms a heterodimer with the related transcription factor MAX. This complex binds to the E box DNA consensus sequence and regulates the transcription of specific target genes. Amplification of this gene is frequently observed in numerous human cancers. Translocations involving this gene are associated with Burkitt lymphoma and multiple myeloma in human patients. There is evidence to show that translation initiates both from an upstream, in-frame non-AUG (CUG) and a downstream AUG start site, resulting in the production of two isoforms with distinct N-termini. |
| NFKB2 | Nuclear Factor Kappa B Subunit 2; Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 2 (P49/P100); Lymphocyte Translocation Chromosome 10 Protein; DNA-Binding Factor KBF2; Oncogene Lyt-10; H2TF1; LYT10; Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 2; Nuclear Factor Of Kappa Light Chain Gene Enhancer In B Cells 2; Nuclear Factor NF-Kappa-B P100 Subunit; Nuclear Factor NF-Kappa-B P52 Subunit | 4791 | Q00653 | This gene encodes a subunit of the transcription factor complex nuclear factor-kappa-B (NFkB). The NFkB complex is expressed in numerous cell types and functions as a central activator of genes involved in inflammation and immune function. The protein encoded by this gene can function as both a transcriptional activator or repressor depending on its dimerization partner. The p100 full-length protein is co-translationally processed into a p52 active form. Chromosomal rearrangements and translocations of this locus have been observed in B cell lymphomas, some of which may result in the formation of fusion proteins. There is a pseudogene for this gene on chromosome 18. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2013] |
| PIAS3 | PIAS3; protein inhibitor of activated STAT, 3; E3 SUMO-protein ligase PIAS3; FLJ14651; zinc finger; MIZ type containing 5; ZMIZ5; E3 SUMO protein ligase PIAS 3; E3 SUMO protein ligase PIAS3; E3 SUMO-protein ligase PIAS3; FLJ14651; OTTHUMP00000015586; OTTHUMP00000015587; PIAS 3; Pias3; PIAS3 protein; PIAS3_HUMAN; Protein inhibitor of activated STAT 3; Protein inhibitor of activated STAT protein 3; Protein inhibitor of activated STAT3; Zinc finger MIZ type containing 5; ZMIZ 5; ZMIZ5; OTTHUMP00000015586; OTTHUMP00000015587; zinc finger, MIZ-type containing 5; protein inhibitor of activated STAT protein 3; | 10401 | Q9Y6X2 | This gene encodes a member of the PIAS [protein inhibitor of activated STAT (signal transducer and activator of transcription)] family of transcriptional modulators. The protein functions as a SUMO (small ubiquitin-like modifier)-E3 ligase which catalyzes the covalent attachment of a SUMO protein to specific target substrates. It directly binds to several transcription factors and either blocks or enhances their activity. Alternatively spliced transcript variants of this gene have been identified, but the full-length nature of some of these variants has not been determined. [provided by RefSeq, Jul 2008] |
| REL | REL Proto-Oncogene, NF-KB Subunit; V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog; Oncogene REL, Avian Reticuloendotheliosis; Proto-Oncogene C-Rel; C-Rel | 5966 | Q04864 | This gene encodes a protein that belongs to the Rel homology domain/immunoglobulin-like fold, plexin, transcription factor (RHD/IPT) family. Members of this family regulate genes involved in apoptosis, inflammation, the immune response, and oncogenic processes. This proto-oncogene plays a role in the survival and proliferation of B lymphocytes. Mutation or amplification of this gene is associated with B-cell lymphomas, including Hodgkin's lymphoma. Single nucleotide polymorphisms in this gene are associated with susceptibility to ulcerative colitis and rheumatoid arthritis. Alternative splicing results in multiple transcript variants encoding different isoforms. |
| RELA | RELA Proto-Oncogene, NF-KB Subunit; Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells 3; V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog A; Nuclear Factor NF-Kappa-B P65 Subunit; NFKB3; V-Rel Reticuloendotheliosis Viral Oncogene Homolog A | 5970 | Q04206 | NF-kappa-B is a ubiquitous transcription factor involved in several biological processes. It is held in the cytoplasm in an inactive state by specific inhibitors. Upon degradation of the inhibitor, NF-kappa-B moves to the nucleus and activates transcription of specific genes. NF-kappa-B is composed of NFKB1 or NFKB2 bound to either REL, RELA, or RELB. The most abundant form of NF-kappa-B is NFKB1 complexed with the product of this gene, RELA. Four transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2011] |
| RELB | IREL; I-REL; IMD53; REL-B | 5971 | Q01201 | Enables RNA polymerase II cis-regulatory region sequence-specific DNA binding activity and protein kinase binding activity. Involved in lymphocyte differentiation and negative regulation of interferon-beta production. Located in cytosol and nucleoplasm. Part of chromatin; nucleus; and transcription repressor complex. Colocalizes with centrosome. Implicated in breast cancer and immunodeficiency 53. Biomarker of breast cancer and transitional cell carcinoma. |
| STAT1 | CANDF7; IMD31A; IMD31B; IMD31C; ISGF-3; STAT91 | 6772 | P42224 | The protein encoded by this gene is a member of the STAT protein family. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. The protein encoded by this gene can be activated by various ligands including interferon-alpha, interferon-gamma, EGF, PDGF and IL6. This protein mediates the expression of a variety of genes, which is thought to be important for cell viability in response to different cell stimuli and pathogens. The protein plays an important role in immune responses to viral, fungal and mycobacterial pathogens. Mutations in this gene are associated with Immunodeficiency 31B, 31A, and 31C. |
| STAT2 | AW496480; 1600010G07Rik | 6773 | P52630 | Involved in negative regulation of type I interferon-mediated signaling pathway and type I interferon signaling pathway. Located in cytoplasm and nucleus. Is expressed in several structures, including central nervous system; early conceptus; and genitourinary system. |
| STAT3 | Signal transducer and activator of transcription 3; Stat3# | 6774 | P40763 | Signal transducer and activator of transcription 3 (STAT3) is a transcription factor which in humans is encoded by the STAT3 gene.[5] It is a member of the STAT protein family. |
| STAT4 | STAT4; Signal transducer and activator of transcription 4 | 6775 | Q14765 | The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein is essential for mediating responses to IL12 in lymphocytes, and regulating the differentiation of T helper cells. |
| STAT6 | STAT6B; STAT6C; D12S1644; IL-4-STAT | 6778 | P42226 | The protein encoded by this gene is a member of the STAT family of transcription factors. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein plays a central role in exerting IL4 mediated biological responses. It is found to induce the expression of BCL2L1/BCL-X(L), which is responsible for the anti-apoptotic activity of IL4. Knockout studies in mice suggested the roles of this gene in differentiation of T helper 2 (Th2) cells, expression of cell surface markers, and class switch of immunoglobulins. Alternative splicing results in multiple transcript variants. |
| TP53 | Tumor Protein P53; Phosphoprotein P53; Antigen NY-CO-13; P53; Transformation-Related Protein 53; Cellular Tumor Antigen P53; Mutant Tumor Protein 53; Li-Fraumeni Syndrome | 7157 | K7PPA8 | This gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Mutations in this gene are associated with a variety of human cancers, including hereditary cancers such as Li-Fraumeni syndrome. Alternative splicing of this gene and the use of alternate promoters result in multiple transcript variants and isoforms. Additional isoforms have also been shown to result from the use of alternate translation initiation codons from identical transcript variants. |
Tested Data-Supported Products for Targeting Apoptosis Transcription Factors and Regulators
- Rusin, Marek. "The p53 protein–not only the guardian of the genome." Postępy Biochemii (2021). Distributed under Open Access license CC BY 4.0, without modification.
 Loading...
Loading...- Human Anti-C-myc Recombinant Antibody; scFv Fragment (HPAB-1760LY-S(P)) (HPAB-1760LY-S(P))
-
- Species Reactivity: Human
- Type: Human scFv
- Application: ELISA
- AbPlus™ Anti-MAX Magnetic Beads (VS-0724-YC1042) (VS-0724-YC1042)
-
- Target: MAX
- Target Species: Human
- Application: IP, Protein Purification
- AbPlus™ Anti-STAT1 Magnetic Beads (VS-0724-YC962) (VS-0724-YC962)
-
- Target: STAT1
- Target Species: Human, Mouse
- Application: IP, Protein Purification
- AbPlus™ Anti-C1D Magnetic Beads (VS-0724-YC903) (VS-0724-YC903)
-
- Target: C1D
- Target Species: Human
- Application: IP, Protein Purification
- AbPlus™ Anti-STAT6 Magnetic Beads (VS-0724-YC633) (VS-0724-YC633)
-
- Target: STAT6
- Target Species: Human
- Application: IP, Protein Purification
- AbPlus™ Anti-STAT3 Magnetic Beads (CBACN-530) (VS-0424-XY249)
-
- Target: STAT3
- Target Species: Human, Mouse, Rat, Zebrafish
- Application: IP, Protein Purification
- AbPlus™ Anti-STAT1 Magnetic Beads (CBACN-527) (VS-0424-XY248)
-
- Target: STAT1
- Target Species: Human, Mouse
- Application: IP, Protein Purification
- AbPlus™ Anti-JUND Magnetic Beads (CBACN-335) (VS-0424-XY165)
-
- Target: JUND
- Target Species: Human
- Application: IP, Protein Purification
- Mouse Anti-STAT3 Recombinant Antibody (VS3-FY2882) (VS3-FY2882)
-
- Species Reactivity: Human, Mouse, Rat, Monkey, Hamster
- Type: Mouse IgG
- Application: WB
- Rabbit Anti-p53 Recombinant Antibody (VS3-FY2827) (VS3-FY2827)
-
- Species Reactivity: Mouse
- Type: Rabbit IgG
- Application: WB, ICC, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG
- Application: WB, IHC-P, IF, ELISA
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC-P, ICC, IF, IP
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC-P
- Mouse Anti-E2F1 Recombinant Antibody (VS3-FY2693) (VS3-FY2693)
-
- Species Reactivity: Human, Mouse, Rat
- Type: Mouse IgG
- Application: WB, ICC, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC-P, ICC, IF, IP, FC
- Rabbit Anti-MAF Recombinant Antibody (clone R04-6K7) (VS3-XY3423)
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: IHC-P
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF
-
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: WB
- Rabbit Anti-REL Recombinant Antibody (clone R03-3D5) (VS3-FY1988)
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, IP
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, IHC-P, IP
- Mouse Anti-IRF6 Recombinant Antibody (clone ZN106) (VS3-FY1584)
-
- Species Reactivity: Human
- Type: Mouse IgG2b
- Application: IHC-P
- Recombinant Mouse Anti-STAT6 Antibody (VS-0923-FY193) (VS-0923-FY193)
-
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: ELISA, WB
- Recombinant Mouse Anti-STAT6 Antibody (VS-0923-FY192) (VS-0923-FY192)
-
- Species Reactivity: Human
- Type: Mouse IgG2a
- Application: ELISA, WB
- Recombinant Mouse Anti-c-Myc Antibody (VS-0923-FY160) (VS-0923-FY160)
-
- Type: Mouse IgG1
- Application: ELISA, WB, IP, IF
- Recombinant Mouse Anti-STAT3 Antibody (VS-0923-FY137) (VS-0923-FY137)
-
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: ELISA, WB
-
- Species Reactivity: Human, Mouse, Rat, Hamster
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, IHC-P, IP
-
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: WB, IHC-P
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ICC, IF, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC-P
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: WB
-
- Derivation: Rabbit
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, IHC, IF, FC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, IHC, IF
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC-F, IHC-P, ICC, IF, IP
- Rabbit Anti-STAT1 Polyclonal Antibody (VS3-WK1565)
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ELISA
- Mouse Anti-STAT3 Recombinant Antibody (VS3-WK1364) (VS3-WK1364)
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG
- Application: WB, IHC-P
-
- Derivation: Rabbit
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP, FC
-
- Derivation: Rabbit
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, FC
-
- Derivation: Rabbit
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: WB, IP, FC
-
- Derivation: Rabbit
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP
- Mouse Anti-RELB Recombinant Antibody (VS3-WK841) (VS3-WK841)
-
- Derivation: Mouse
- Species Reactivity: Human, Mouse
- Type: Mouse IgG
- Application: WB
-
- Derivation: Rabbit
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP, FC
-
- Species Reactivity: Human
- Application: WB
- Mouse Anti-RELB Recombinant Antibody (clone 2E4B1) (VS3-XY1367)
-
- Species Reactivity: Human
- Type: Mouse IgG2a
- Application: ICC, FC
- Mouse Anti-MSX1 Recombinant Antibody (clone 5D11) (VS3-XY1120)
-
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: ELISA, WB
-
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: ELISA, WB, IHC
- Rabbit Anti-FOSB Recombinant Antibody (VS3-CJ1035) (VS3-CJ1035)
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP
-
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP, FC
-
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: WB
-
- Species Reactivity: Human, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC, IP
-
- Species Reactivity: Human, Mouse, Rat
- Type: Mouse IgG
- Application: WB, IHC
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ICC, IF, IP
- Mouse Anti-ETS1 Recombinant Antibody (VS3-CJ658) (VS3-CJ658)
-
- Species Reactivity: Human
- Type: Mouse IgG2b, κ
- Application: WB, FC
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, IHC
-
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IF, IHC
-
- Species Reactivity: Human
- Type: Mouse IgG
- Application: WB, ELISA
-
- Derivation: Rabbit
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: WB, ELISA
-
- Derivation: Rabbit
- Species Reactivity: Human, Mouse, Rat
- Type: Rabbit IgG
- Application: WB, ICC, IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Mouse
- Type: Mouse IgG2b
- Application: WB, ICC, IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Mouse, Rat, Monkey
- Type: Mouse IgG1
- Application: WB, IHC, ICC, FC, ELISA
-
- Derivation: Mouse
- Species Reactivity: Human, Mouse, Monkey
- Type: Mouse IgG1
- Application: WB, IHC, ICC, FC, ELISA
- Mouse Anti-STAT3 Recombinant Antibody (clone 7G3H4) (VS3-QX1067)
-
- Derivation: Mouse
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: WB, IHC, ICC, FC, ELISA
-
- Derivation: Mouse
- Species Reactivity: Human, Mouse, Rat, Monkey, Hamster
- Type: Mouse IgG1
- Application: WB
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG1
- Application: WB, IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG1
- Application: WB, IHC
-
- Derivation: Mouse
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: IHC
-
- Derivation: Mouse
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: WB
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG1
- Application: IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG1
- Application: IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Rat, Mouse
- Type: Mouse IgG1
- Application: IHC
-
- Derivation: Mouse
- Species Reactivity: Human, Monkey
- Type: Mouse IgG2a
- Application: WB, IP
-
- Derivation: Mouse
- Species Reactivity: Human
- Type: Mouse IgG2b
- Application: WB, ICC
- Mouse Anti-ETS1 Recombinant Antibody (clone 8A8) (VS3-QX392)
-
- Derivation: Mouse
- Species Reactivity: Human
- Type: Mouse IgG1
- Application: WB, IHC, ICC, FC, ELISA
-
- Derivation: Mouse
- Species Reactivity: Human, Rat
- Type: Mouse IgG2b
- Application: WB, IF, IP
-
- Species Reactivity: Human
- Type: Mouse IgG2b
- Application: IF, IP, FC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, FC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC, IP
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC, IF
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC, IF
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, WB, IHC
-
- Species Reactivity: Human, Rat
- Type: Rabbit IgG
- Application: ELISA, WB, IHC, IF, FC
-
- Species Reactivity: Human, Mouse
- Type: Rabbit IgG
- Application: ELISA, WB, IHC
-
- Species Reactivity: Human
- Type: Rabbit IgG
- Application: ELISA, IHC
-
- Species Reactivity: Human, Mouse, Rat, Monkey
- Type: Mouse IgG
- Application: WB, ELISA, IF
-
- Species Reactivity: Human, Mouse, Rat, Monkey, Hamster
- Type: Mouse IgG
- Application: WB
-
- Species Reactivity: Human, Mouse, Monkey
- Type: Mouse IgG
- Application: WB, ELISA, IHC, IF
Our customer service representatives are available 24 hours a day, from Monday to Sunday. Contact Us
Can't find the products you're looking for? Try to filter in the left sidebar.Filter By Tag
For Research Use Only. Not For Clinical Use.









































































