Anti-P. falciparum AMA1 Recombinant Antibody (MRO-191CT)
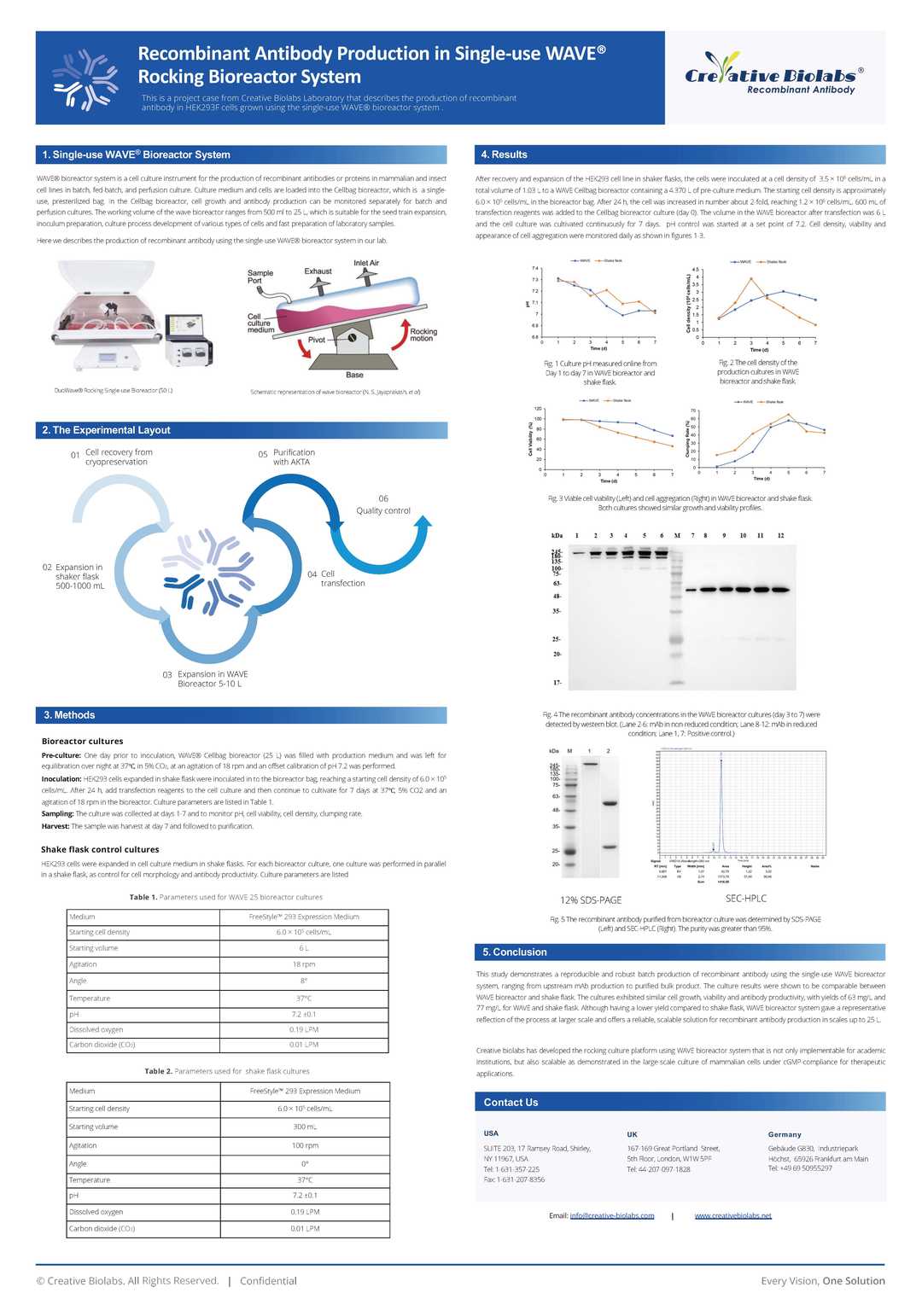
CAT#: MRO-191CT
Recombinant shark antibody specifically binds to Plasmodium falciparum AMA1, expressed in Chinese Hamster Ovary cells(CHO).



Specifications
- Host Species
- Shark
- Antibody Isotype
- IgNAR
- Species Reactivity
- P. falciparum
- Clone
- MRO-191CT
- Applications
- ELISA, SPR, Inhib
Applications
- Application Notes
- The antibody was validated for ELISA, Surface plasmon resonance, Inhibition. For details, refer to Published Data.
Target
Customer Review
There are currently no Customer reviews or questions for MRO-191CT. Click the button above to contact us or submit your feedback about this product.
Submit Your Publication
Published with our product? Submit your paper and receive a 10% discount on your next order! Share your research to earn exclusive rewards.
Downloadable Resources
Download resources about recombinant antibody development and antibody engineering to boost your research.
Product Notes
This is a product of Creative Biolabs' Hi-Affi™ recombinant antibody portfolio, which has several benefits including:
• Increased sensitivity
• Confirmed specificity
• High repeatability
• Excellent batch-to-batch consistency
• Sustainable supply
• Animal-free production
See more details about Hi-Affi™ recombinant antibody benefits.
Datasheet
MSDS
COA
Certificate of Analysis LookupTo download a Certificate of Analysis, please enter a lot number in the search box below. Note: Certificate of Analysis not available for kit components.
See other products for "AMA1"
Select a product category from the dropdown menu below to view related products.
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PABL-293 | Recombinant Rat Anti-P. Knowlesi AMA-1 Antibody (R31C2 ) | WB, BL, FuncS | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PSBL-293 | Recombinant Rat Anti-P. Knowlesi AMA-1 Antibody scFv Fragment (R31C2 ) | WB, BL, FuncS | scFv |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PFBL-293 | Recombinant Rat Anti-P. Knowlesi AMA-1 Antibody Fab Fragment (R31C2 ) | WB, BL, FuncS | Fab |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PABW-097 | Recombinant Mouse Anti-P. falciparum AMA1 Antibody (1F9) | ELISA, BL | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PABZ-098 | Recombinant Mouse Anti-P. vivax AMA-1 Antibody (F8.12.19) | IF | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PABZ-148 | Recombinant Mouse Anti-P. vivax AMA1 Antibody (F8.12.19) | WB | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PABL-397 | Human Anti-AMA1 Recombinant Antibody (PABL-397) | WB, ELISA, FuncS | Human IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PFBW-097 | Recombinant Mouse Anti-P. falciparum AMA1 Antibody Fab Fragment (1F9) | ELISA, BL | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PFBZ-098 | Recombinant Mouse Anti-P. vivax AMA-1 Antibody Fab Fragment (F8.12.19) | IF | IgG |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PFBZ-148 | Recombinant Mouse Anti-P. vivax AMA1 Antibody Fab Fragment (F8.12.19) | WB | Fab |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PFBL-396 | Human Anti-AMA1 Recombinant Antibody; Fab Fragment (PFBL-396) | WB, ELISA, FuncS | Human Fab |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PSBW-097 | Recombinant Mouse Anti-P. falciparum AMA1 Antibody scFv Fragment (1F9) | ELISA, BL | scFv |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PSBZ-098 | Recombinant Mouse Anti-P. vivax AMA-1 Antibody scFv Fragment (F8.12.19) | IF | scFv |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PSBZ-148 | Recombinant Mouse Anti-P. vivax AMA1 Antibody scFv Fragment (F8.12.19) | WB | scFv |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| PSBL-396 | Human Anti-AMA1 Recombinant Antibody; scFv Fragment (PSBL-396) | WB, ELISA, FuncS | Human scFv |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| MRO-190CT | Anti-P. falciparum AMA1 Recombinant Antibody (MRO-190CT) | ELISA, SPR |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| MRO-192CT | Anti-P. falciparum AMA1 Recombinant Antibody (MRO-192CT) | ELISA, SPR |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| MRO-193CT | Anti-P. falciparum AMA1 Recombinant Antibody (MRO-193CT) | ELISA, SPR, Inhib |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| MRO-194CT | Anti-P. falciparum AMA1 Recombinant Antibody (MRO-194CT) | ELISA, SPR, Inhib |
| CAT | Product Name | Application | Type |
|---|---|---|---|
| MRO-190CT-S(P) | Anti-P. falciparum AMA1 Recombinant Antibody scFv Fragment (MRO-190CT-S(P)) | ELISA |
Popular Products

Application: FC, IP, ELISA, Neut, FuncS, IF, IHC

Application: ELISA, IP, FC, FuncS, Neut, IF, IHC

Application: IF, IP, Neut, FuncS, ELISA, FC, WB

Application: WB, FC, IP, ELISA, Neut, FuncS, IF

Application: WB, FC, IP, ELISA, Neut, FuncS, IF

Application: FC, IHC, FuncS, Inhib, Cyt

Application: ELISA, FC, IP, FuncS, IF, Neut, ICC

Application: ELISA, FC, IP, FuncS, IF, Neut, ICC

Application: ELISA, FC, WB, FuncS

Application: ELISA, FC

Application: ELISA, FuncS

Application: ELISA, FC, Neut, Inhib

Application: Neut, ELISA, IF, IP, FuncS, FC

Application: ELISA, Inhib, FuncS
For research use only. Not intended for any clinical use. No products from Creative Biolabs may be resold, modified for resale or used to manufacture commercial products without prior written approval from Creative Biolabs.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.