Methylated DNA Immunoprecipitation Protocol & Troubleshooting
DNA methylation is a major epigenetic modification that occurs primarily at the C5 position of the cytosine ring. Methylated DNA immunoprecipitation (MeDIP) is a technique that captures and enriches methylated DNA using 5mC-specific antibodies. This technique enables the isolation of methylated DNA fragments for subsequent analysis.
We present a standard MeDIP method. In this protocol, antibodies against 5mC are used to immunoprecipitate methylated DNA. And we can also provide highly specific antibodies and the necessary solutions and reagents. The rapid and streamlined protocol helps you save valuable time.
Solutions and Reagents
| Stages | Solutions and Reagents |
| Sample Preparation | Digestion buffer, nucleic acid extraction solution, DNA purify cation kit, buffer TE, IP buffer |
| Immunoprecipitation | IP buffer, anti-5mC antibody, dilution buffer, digestion buffer, wash buffer, binding buffer, elution buffer |
Methylated DNA Immunoprecipitation Procedure
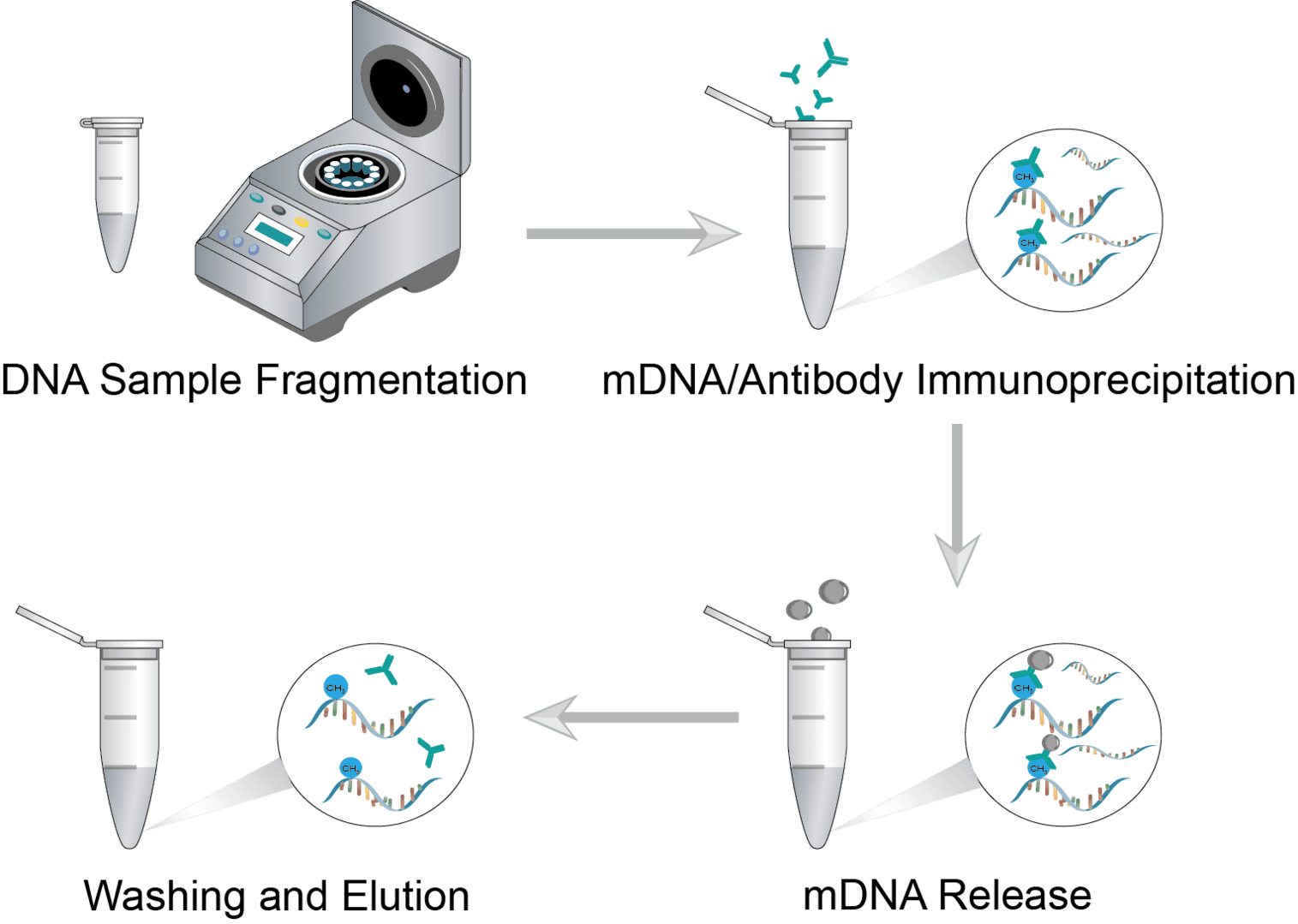
Collect the cells and add digestion buffer for cell lysis. Then extract the nucleic acids from the sample and perform DNA purification. Before starting the assay, genomic DNA should be fragmented to produce fragments between 200 and 600 base pairs in size. Transfer the quantitative genomic DNA sample into a microcentrifuge tube and adjust the final volume. Perform sonication on ice using a sonicator. If desired, the sonicated DNA can be verified with agar gel analysis.
To further increase the binding affinity of the antibody, the DNA fragment is denatured to produce single-stranded DNA. After denaturation, place the DNA on ice immediately and then add IP buffer to mix thoroughly. Then incubate the DNA with the appropriate antibody dilution.
Add the methylated DNA/antibody complex to the beads and incubate. At the end of the incubation, centrifuge the bead-bound methylated DNA/antibody complex and discard the supernatant. Add IP buffer and wash several times. If using magnetic beads, no centrifugation is required after incubation with the DNA/antibody complex. Collect the beads directly using the magnetic rack and repeat the wash.
Resuspend the collected magnetic beads in the elution buffer and heat to incubate. After incubation is complete, centrifuge the supernatant and repeat the elution step several times, combining the eluates in a single tube. If using magnetic beads, place the suspension on a magnetic separation holder and repeat the elution step as above to collect the supernatant.
Troubleshooting
After MeDIP, DNA methylation can be analyzed using a variety of downstream applications. To ensure that your enriched mDNA samples are rich and accurate, you can refer to our troubleshooting tips below to note relevant experimental factors.
Little or no enrichment of methylated DNA
- Sample causes. Insufficient starting DNA may result in less final enrichment of methylated DNA. You need to make sure that the amount of starting DNA is sufficient.
- DNA Shearing causes. Inadequate or excessive sonication can result in DNA that is not of the required size. Follow the protocol instructions to obtain DNA of appropriate size and keep the sample on the ice during sonication. If you use restriction digestion for DNA shearing it is also possible. However, this requires that the DNA must be well purified and different enzymes may need to be used for different motifs. We recommend treating the sample at 65°C after restriction digestion to inactivate the remaining enzymes.
- Methylated DNA release causes. Check and make sure that the buffer solution is not contaminated, and that the wash time is too long can also cause a reduced signal. Please follow the guidelines for proper temperature and time in the relevant protocol.
- Antibody causes. First, you need to know that the 5-methylcytosine antibody can only be used for single-stranded DNA. It is critical to place the DNA on ice immediately after heating to 95°C to prevent re-annealing. Also, if you use a different DNA concentration then you need to change the antibody concentration as well. A lack of antibodies will reduce the recovery of methylated DNA, while an excess of antibodies will reduce the specificity of the IP.
- DNA purification column causes. There are many brands and types of DNA purification columns on the market. So, it is necessary for you to confirm whether the purification column is suitable for capturing single-stranded DNA. It can be used after verification.
- Washing causes. Perform appropriate washes as specified in the protocol, ensuring that each wash step is adequate. Especially, the step of eluting the DNA sample with the column should be performed rigorously.
If you are planning to conduct MeDIP experiments, please visit our page and perform a search to view the published MeDIP-related products and services using our technical filters.
For research use only. Not intended for any clinical use.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.



