Surface Plasmon Resonance Protocol & Troubleshooting
Surface plasmon resonance (SPR) is an optical sensing method for label-free detection of biomolecular interactions. The method is based on immobilization of a ligand on the sensor surface and reinjection of the analyses onto the surface containing the ligand. After injection, binding is monitored by changes in the refractive index of the medium close to the sensor surface. It is commonly used for detailed and quantitative studies of protein-protein interactions as well as kinetics.
We introduce the basic concepts of SPR and how it can be used to analyze and visualize biomolecular interactions. We provide SPR specific experimental protocols and troubleshooting for drug development, life science research, and other fields.
Solutions and Reagents
| Stages | Solutions and Reagents |
| Preparation | Diluting solution, ligand, analyte |
| Immobilization | Running buffer, activation buffer, immobilization buffer, stabilization buffer, |
Surface Plasmon Resonance Procedure
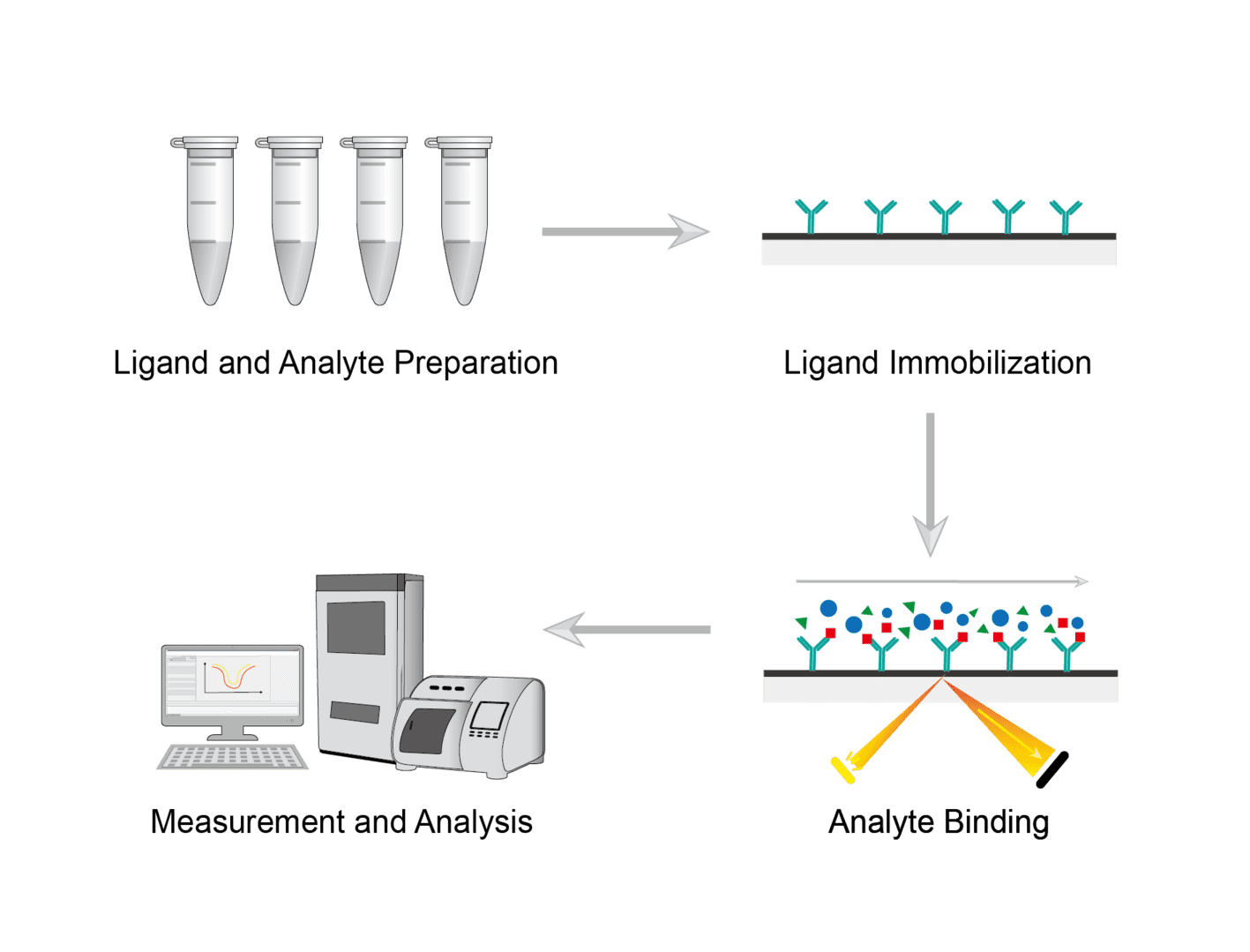
Express and purify ligands and analytes. Check the purity and stability of the protein. Prepare various materials and buffers for later use.
Select a suitable sensor chip. Perform conditioning and activation of the sensor chip to form reactive groups on the sensor chip surface. Which sensor chip to use depends on the degree of immobilization of the desired ligand and the specific application. Determine the optimal pH of the immobilization buffer and the concentration of the ligand. Inject the ligand protein with the desired injection parameters to achieve immobilization.
Measure the concentration of the analyte. Dilute the analyte using the running buffer. Place the sample in the selected position and insert it into the sample holder. Inject the analyte for binding analysis.
It is possible to select different modes of analysis on the instrument. Select binding analysis and direct binding to study protein binding. Select kinetic analysis and choose mass transfer to run kinetic analysis and binding experiments. Perform data analysis after data is obtained.
Troubleshooting
Inactive targets
- Sample causes. Your target protein may have been inactivated. We recommend that you check your sample proteins before performing spiked analysis. Ensure the quality of the target protein.
- Sensor chip causes. Another possibility is that it is not due to your protein itself. Rather, the surface of the sensor chip may show low binding activity for your analyte. You can try coupling the target to the slide in a different way to improve binding.
Non-specific binding
- Analyte causes. Analytes may not only bind to the SPR surface with respect to the target, but may also bind non-specifically. We recommend minimizing non-specific binding by adding additives such as surfactants or bovine serum albumin (BSA) to the run buffer. If appropriate, you can also add dextran or polyethylene glycol (PEG).
- Ligand causes. Another way to avoid non-specific binding is to couple a ligand compound that does not bind to the analyte on the reference.
- Sensor chip causes. You can sometimes change the type of sensor chip you have as well.
Negative binding signals
- Analyte concentration causes. We recommend testing the suitability of your reference channel. And you can improve the signal by injecting the highest concentration of analyte on the surface.
- Buffer mismatch causes. You see a binding signal in the SPR assay, but it is stronger than the binding to the target. This may be caused by buffer mismatch or other non-specific interactions. There are many approaches you can take to address these causes.
Regeneration problems in SPR instrument
- Solution causes. You need to determine the appropriate solution to regenerate the sensor surface. This way the chip can be used repeatedly for multiple injections of analytes. Many different solutions can be chosen for testing, including acidic solutions, alkaline solutions, or high salt solutions. In addition, the addition of 10% glycerol is helpful for target stability.
We will be happy to discuss the details of your anticipated surface plasmon resonance and develop an experimental approach based on your requirements. Please review the SRP service on our website and contact Creative Biolabs.
Products with Tested Data
At Creative Biolabs, we are dedicated to providing high-quality antibodies for various research applications. Each product in our extensive range has been rigorously tested to ensure superior reliability and efficacy. To showcase the performance of our antibodies, we have conducted numerous experiments using Surface plasmon resonance (SPR) assay. Below, you will find a table listing a selection of our antibody products along with images from these experiments, demonstrating their proven reliability.
| Product Name | Catalog Number | Target | Image | Description |
|---|---|---|---|---|
| Human Anti-ERBB2 Rcombinant Antibody (TP-083CL) | TP-083CL | IGHG1 |

|
SPR assays of TP-083CL. The affinity of Trastuzumab (TP-083CL) was determined by SPR. Captured TP-083CL on CM5 Chip via Anti-Human IgG (Fc) can bind Human ERBB2 protein with an affinity constant of 1.45 nM as determined in a SPR assay (Biacore 8K). |
| Anti-ERBB2 Recombinant Antibody (Margetuximab) | TAB-761 | ERBB2 |

|
SPR assays of TAB-761. The affinity of Anti-ERBB2 Therapeutic Antibody (Margetuximab) (TAB-761) was determined by SPR. Captured TAB-761 on CM5 Chip via Anti-Human IgG (Fc) can bind Human ERBB2 protein with an affinity constant of 1.21 nM as determined in a SPR assay (Biacore 8K). |
| Anti-Human ERBB2 Recombinant Antibody (TAB-053) | TAB-053 | ERBB2 |

|
SPR assays of TAB-053. The affinity of Anti-Human ERBB2 Antibody (TAB-053) was determined by SPR. Captured TAB-053 on CM5 Chip via Anti-Human IgG (Fc) can bind Human ERBB2 protein with an affinity constant of 1.88 nM as determined in a SPR assay (Biacore 8K). |
| Anti-Human CD19 Recombinant Antibody scFv Fragment (HD37) | TAB-1620CL-S(P) | CD19 |
-1.png)
|
SPR assays of TAB-1620CL-S(P). Antibody capture Chip principle: Immobilization: Immobilized Anti-Human IgG (Fc) antibody on CM5 chip Capture: capture the Human CD19,Fc tag Analyte: the analyte is TAB-1620CL-S(P) Regeneration: 3M MgCl2 |
| Human Anti-PDCD1 Recombinant Antibody (TAB-H55) | TAB-H55 | PDCD1 |

|
Dilute the antibody ligand (5 μg/mL) and antigen analyte with running buffer. The ligand was injected to sample channel (Fc2) at a flow rate of 10 μL/min to reach a capture level of about 400 RU. The analyte was injected to Fc1-Fc2 of the channel at a flow rate of 30 μL/min for an association phase of the corresponding time. The association and dissociation process were all handling in the running buffer. Repeat 6 cycles of analyte according to analyte concentrations in ascending order. |
| Anti-PDCD1 Recombinant Antibody (TAB-770) | TAB-770 | PDCD1 |

|
Dilute the antibody ligand (5 μg/mL) and antigen analyte with running buffer. The ligand was injected to sample channel (Fc2) at a flow rate of 10 μL/min to reach a capture level of about 400 RU. The analyte was injected to Fc1-Fc2 of the channel at a flow rate of 30 μL/min for an association phase of the corresponding time. The association and dissociation process were all handling in the running buffer. Repeat 6 cycles of analyte according to analyte concentrations in ascending order. |
For research use only. Not intended for any clinical use.
This site is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.



